Note
Go to the end to download the full example code.
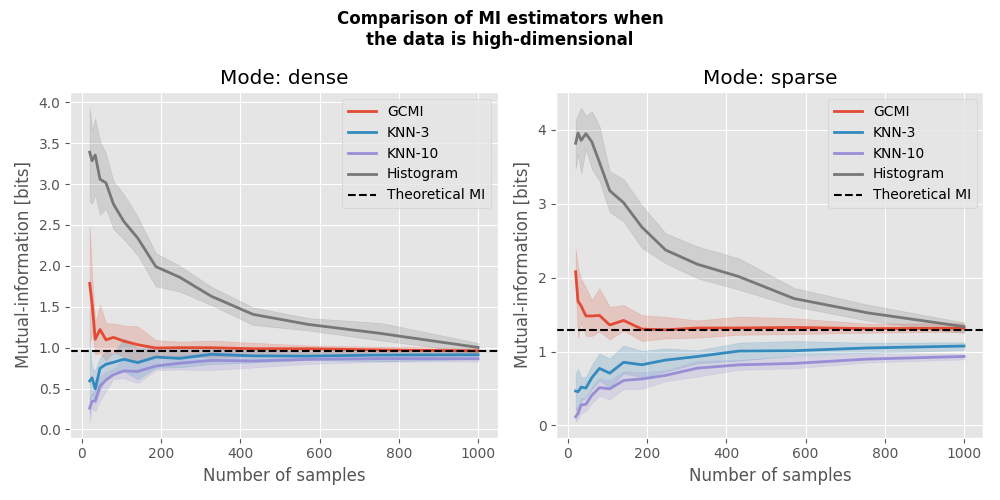
Comparison of MI estimators with high-dimensional data#
In this example, we are going to compare estimators of mutual-information (MI) with high-dimensional data. In particular, the MI between variables sampled from a multinormal distribution can be estimated theoretically. In this this tutorial we are going to:
Simulate data sampled from a “splitted” multivariate normal distribution.
Define estimators of MI.
Compute the MI for a varying number of samples.
See if the computed MI converge towards the theoretical value.
This example is inspired from a similar simulation done by Czyz et al., NeurIPS 2023 [6].
import matplotlib.pyplot as plt
import numpy as np
from hoi.core import get_mi
plt.style.use("ggplot")
Definition of MI estimators#
We are going to use the GCMI (Gaussian Copula Mutual Information), KNN (k Nearest Neighbor) and histogram estimator.
# list of estimators to compare
metrics = {
"GCMI": get_mi("gc", biascorrect=False),
"KNN-3": get_mi("knn", k=3),
"KNN-10": get_mi("knn", k=10),
"Histogram": get_mi("histogram", n_bins=3),
}
# number of samples to simulate data
n_samples = np.geomspace(20, 1000, 15).astype(int)
# number of repetitions to estimate the percentile interval
n_repeat = 10
# plotting function
def plot(mi, mi_theoric, ax):
"""Plotting function."""
for n_m, metric_name in enumerate(mi.keys()):
# get the entropies
x = mi[metric_name]
# get the color
color = f"C{n_m}"
# estimate lower and upper bounds of the [5, 95]th percentile interval
x_low, x_high = np.percentile(x, [5, 95], axis=0)
# plot the MI as a function of the number of samples and interval
ax.plot(n_samples, x.mean(0), color=color, lw=2, label=metric_name)
ax.fill_between(n_samples, x_low, x_high, color=color, alpha=0.2)
# plot the theoretical value
ax.axhline(mi_theoric, linestyle="--", color="k", label="Theoretical MI")
ax.legend()
ax.set_xlabel("Number of samples")
ax.set_ylabel("Mutual-information [bits]")
MI of data sampled from splitted multinormal distribution#
Given two variables \(X \sim \mathcal{N}(\vec{\mu_{x}}, \Sigma_{x})\) and \(Y \sim \mathcal{N}(\vec{\mu_{y}}, \Sigma_{y})\), linked by a covariance \(C\) the theoretical MI in bits is defined by :
# function for creating the covariance matrix with differnt modes
def create_cov_matrix(n_dims, cov, mode="dense", k=None):
"""Create a covariance matrix."""
# variance of x and y, for each dimension, 1
cov_matrix = np.eye(2 * n_dims)
if mode == "dense":
# all dimensions, but diagonal, with covariance cov
cov_matrix += cov
cov_matrix[np.diag_indices(2 * n_dims)] = 1
elif mode == "sparse":
# only pairs xi, yi with i < k have covariance cov
k = k if k is not None else n_dims
for i in range(n_dims):
if i < k:
cov_matrix[i, i + n_dims] = cov
cov_matrix[i + n_dims, i] = cov
return cov_matrix
def compute_true_mi(cov_matrix):
"""Compute the true MI (bits)."""
n_dims = cov_matrix.shape[0] // 2
det_x = np.linalg.det(cov_matrix[n_dims:, n_dims:])
det_y = np.linalg.det(cov_matrix[:n_dims, :n_dims])
det_xy = np.linalg.det(cov_matrix)
return 0.5 * np.log2(det_x * det_y / det_xy)
# number of dimensions per variable
n_dims = 4
# mean
mu = [0.0] * n_dims * 2
# covariance
covariance = 0.6
# modes for the covariance matrix:
# - dense: off diagonal elements have specified covariance
# - sparse: only pairs xi, yi with i < k have specified covariance
modes = ["dense", "sparse"]
# number of pairs with specified covariance
k = n_dims
fig = plt.figure(figsize=(10, 5))
# compute mi using various metrics
mi = {k: np.zeros((n_repeat, len(n_samples))) for k in metrics.keys()}
for i, mode in enumerate(modes):
cov_matrix = create_cov_matrix(n_dims, covariance, mode=mode)
# define the theoretic MI
mi_theoric = compute_true_mi(cov_matrix)
ax = fig.add_subplot(1, 2, i + 1)
for n_s, s in enumerate(n_samples):
for n_r in range(n_repeat):
# generate samples from joint gaussian distribution
fx = np.random.multivariate_normal(mu, cov_matrix, s)
for metric, fcn in metrics.items():
# extract x and y
x = fx[:, :n_dims].T
y = fx[:, n_dims:].T
# compute mi
mi[metric][n_r, n_s] = fcn(x, y)
# plot the results
plot(mi, mi_theoric, ax)
ax.title.set_text(f"Mode: {mode}")
fig.suptitle(
"Comparison of MI estimators when\nthe data is high-dimensional",
fontweight="bold",
)
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 29.929 seconds)
