frites.conn.conn_ccf#
- frites.conn.conn_ccf(data, times=None, roi=None, normalized=True, n_jobs=1, times_as_sample=True, sfreq=None, verbose=None, **kw_links)[source]#
Single trial Cross-Correlation Function.
This function computes the pairwise Cross Correlation (CCF) at the single trial level. This can be particulary usefull to find whether there are temporal delays between times series.
- Parameters:
- datanumpy:array_like
Electrophysiological data. Several input types are supported :
Standard NumPy arrays of shape (n_epochs, n_roi, n_times)
mne.Epochs
xarray.DataArray of shape (n_epochs, n_roi, n_times)
- timesnumpy:array_like |
python:None Time vector array of shape (n_times,). If the input is an xarray, the name of the time dimension can be provided
- roinumpy:array_like |
python:None ROI names of a single subject. If the input is an xarray, the name of the ROI dimension can be provided
- normalizedbool |
python:True Z-score normalization of the data. By default, it set to true.
- times_as_samplebool |
python:True Specify whether the time dimension of the cross-correlation output should be described using the time unit of the input data or in samples. By default, samples are used to describe lags between time-series.
- n_jobs
python:int| 1 Number of jobs to use for parallel computing (use -1 to use all jobs). The parallel loop is set at the pair level.
- sfreq
python:float|python:None The sampling frequency.
- kw_links
python:dict| {} Additional arguments for selecting links to compute are passed to the function
frites.conn.conn_links()
- Returns:
- ccfnumpy:array_like
The Cross-Correlation array of shape (n_epochs, n_pairs, n_times). When the peak of correlation occurs at a negative time it means that the target has to be moved toward the source. On the contrary, if the peak occurs at positive time it means that the target is moved away of the source.
See also
Examples using frites.conn.conn_ccf#
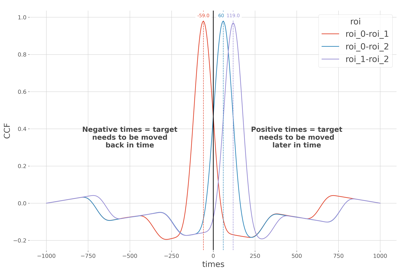
Lag estimation between delayed times-series using the cross-correlation