frites.estimator.GCMIEstimator#
- class frites.estimator.GCMIEstimator(mi_type='cc', copnorm=True, biascorrect=True, demeaned=False, tensor=True, gpu=False, verbose=None)[source]#
Gaussian Copula Mutual-Information estimator.
- Parameters:
- mi_type{‘cc’, ‘cd’, ‘ccd’, ‘ccc’}
Mutual information type (default : ‘cc’) :
‘cc’ : MI between two continuous variables
‘cd’ : MI between a continuous and a discret variables
‘ccd’ : MI between two continuous variables conditioned by a third discret one
‘ccc’ : MI between two continuous variables conditioned by a third continuous one
- copnormbool |
python:True Apply the gaussian-copula rank normalization (default : True)
- biascorrectbool |
python:True Specifies whether bias correction should be applied to the estimated MI (default : True)
- demeanedbool |
python:False Specifies whether the input data already has zero mean (true if it has been copula-normalized) (default : False)
- tensorbool |
python:True Specify the implementation of the GCMI, either tensor-based (Nd) or vector-based (1d). Usually, tensor-based implementation is faster but also requires more RAM (default : True)
- gpubool |
python:False Specify whether the mutual-information has to be computed on CPU (gpu=False) or on GPU (gpu=True) (default : False)
Methods
estimate(x, y[, z, categories])Estimate the (possibly conditional) mutual-information.
Get the function to execute according to the input parameters.
- estimate(x, y, z=None, categories=None)[source]#
Estimate the (possibly conditional) mutual-information.
This method is made for computing the mutual-information on 3D variables (i.e (n_var, n_mv, n_samples)) where n_var is an additional dimension (e.g times, times x freqs etc.), n_mv is a multivariate axis and n_samples the number of samples. When computing MI, both the multivariate and samples axes are reduced.
- Parameters:
- xnumpy:array_like
Array of shape (n_var, n_mv, n_samples). If x has more than three dimensions, it’s going to be internally reshaped.
- ynumpy:array_like
Array with a shape that depends on the type of MI (mi_type) :
If mi_type is ‘cc’ or ‘ccd’, y should be an array with the same shape as x
If mi_type is ‘cd’, y should be a row vector of shape (n_samples,)
- znumpy:array_like |
python:None Array for conditional mutual-information. The shape is going to depend on the type of MI (mi_type) :
If mi_type is ‘ccd’, z should be a row vector of shape (n_samples,)
If mi_type is ‘ccc’, z should have the same shape as x and y
- categoriesnumpy:array_like |
python:None Row vector of categories. This vector should have a shape of (n_samples,) and should contains integers describing the category of each sample. If categories are provided, the copnorm is going to be performed per categories.
- Returns:
- minumpy:array_like
Array of (possibly conditional) mutual-information of shape (n_categories, n_var). If categories is None when computing MI, n_categories is going to be one.
- get_function()[source]#
Get the function to execute according to the input parameters.
This can be particularly useful when computing MI in parallel as it avoids to pickle the whole estimator and therefore, leading to faster computations.
The returned function has the following signature :
fcn(x, y, z=None, categories=None)
and return an array of shape (n_categories, n_var).
Examples using frites.estimator.GCMIEstimator#
Trial-resampling: correcting for unbalanced designs
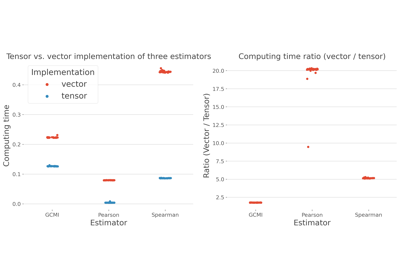
Comparison between tensor and vector based computations