frites.dataset.DatasetEphy#
- class frites.dataset.DatasetEphy(x, y=None, z=None, roi=None, agg_ch=True, times=None, multivariate=False, nb_min_suj=False, attrs=None, verbose=None)[source]#
Multi-subjects electrophysiological data container.
This class is a container used to represent the neurophysiological data coming from multiple subjects. In addition to passing the data, this container is also going to need the anatomical information such as the task-related variables (continuous or discret). Then, the created object can be used to compute the mutual information (MI) between the data and the task variable and group-level.
- Parameters:
- x
python:list List of length (n_subjects,) where each element of the list should be the neural data of single subject. Supported types for each element of the list are the same as in
SubjectEphy- y, z
python:list,sting|python:None List of length (n_subjects,) of continuous or discrete task-related variables per subject. Supported types for each element of the list are the same as in
SubjectEphy- roi
python:list|python:None List of length (n_subjects,) where each element represents the anatomical information of each channel. Supported types for each element of the list are the same as in
SubjectEphy- agg_chbool |
python:True If multiple channels belong to the same ROI, specify whether if the data should be aggregated across channels (True) or if the information per channel have to be take into account (False - conditional mutual information).
- timesnumpy:array_like |
python:None The time vector to use. Supported types are listed in
SubjectEphy- multivariatebool |
python:False If 4d input is provided, this parameter specifies whether this axis should be considered as multi-variate (True) or uni-varariate (False)
- nb_min_suj
python:int|python:None The minimum number of subjects per roi. Roi with n_suj < nb_min_suj are going to be skipped. Use None to force selecting all of the subjects
- attrs
python:dict| {} Dictionary of additional attributes about the data
- x
- Attributes:
Methods
get_connectivity_pairs([as_blocks, ...])Get the connectivity pairs for this dataset.
get_roi_data(roi[, groupby, mi_type, ...])Get the data of a single brain region.
isel(**kwargs)Index-based data slicing.
savgol_filter(h_freq[, edges, verbose])Filter the data using Savitzky-Golay polynomial method.
sel(**kwargs)Coordinate-based data slicing.
- property df_rs#
Pandas DataFrame of cross-subjects anatomical repartition.
- get_connectivity_pairs(as_blocks=False, directed=False, verbose=None)[source]#
Get the connectivity pairs for this dataset.
This method can be used to get the possible connectivity pairs i.e (sources, targets) for directed connectivity (or not). In addition, some pairs are going to be ignored because of a number of subjects to low.
- Parameters:
- directedbool |
python:False Get either directed (True) or non-directed (False) pairs
- directedbool |
- Returns:
- df_conn
pd.DataFrame The table describing the connectivity informations per pair of brain regions
- df_conn_suj
pd.DataFrame The table describing the connectivity informations per subject
- df_conn
- get_roi_data(roi, groupby='subjects', mi_type='cc', copnorm=True, gcrn_per_suj=True)[source]#
Get the data of a single brain region.
- Parameters:
- roi
python:str ROI name to get
- groupby{‘subjects’}
Specify if the data across subjects have to be concatenated
- mi_type{‘cc’, ‘cd’, ‘ccd’}
The type of mutual-information that is then going to be used. This is going to have an influence on how the data are organized and how the copnorm is going to be applied
- copnormbool |
python:True Apply the gaussian copula rank normalization
- gcrn_per_sujbool |
python:True Specify whether the gaussian copula rank normalization have to be applied per subject (True - RFX) or across subjects (False - FFX)
- roi
- Returns:
- da
xr.DataArray The data of the single brain region
- da
- isel(**kwargs)[source]#
Index-based data slicing.
Slice the entire dataset based on indexes.
- Parameters:
- kwargs{} |
python:None Additional inputs are sent to to the method xr.DataArray.isel of the data of each subject.
- kwargs{} |
- Returns:
- inst
instanceofDatasetEphy The sliced object.
- inst
Examples
>>> # define the dataset >>> ds = DatasetEphy(...) >>> # temporal slicing of the data between time points (100, 2500) >>> ds.sel(times=slice(100, 2500))
- property roi_names#
List of brain regions to keep across subjects.
- savgol_filter(h_freq, edges=None, verbose=None)[source]#
Filter the data using Savitzky-Golay polynomial method.
This method is an adaptation of the mne-python one. Note that this smoothing operation is performed inplace to avoid data copy.
- Parameters:
- h_freq
python:float Approximate high cut-off frequency in Hz. Note that this is not an exact cutoff, since Savitzky-Golay filtering is done using polynomial fits instead of FIR/IIR filtering. This parameter is thus used to determine the length of the window over which a 5th-order polynomial smoothing is used.
- edges
python:int,python:float|python:None Edge compensation. Use either an integer to drop a specific number of time points (e.g edges=100 remove 100 time points at the begining and at the end) or a float to drop a period (e.g edges=0.2 drop 200ms at the begining and at the end)
- h_freq
- Returns:
- inst
instanceofDatasetEphy The object with the filtering applied.
- inst
Notes
- For Savitzky-Golay low-pass approximation, see:
Examples using
savgol_filter:
- sel(**kwargs)[source]#
Coordinate-based data slicing.
Slice the entire dataset based on the coordinates values.
- Parameters:
- kwargs{} |
python:None Additional inputs are sent to to the method xr.DataArray.sel of the data of each subject.
- kwargs{} |
- Returns:
- inst
instanceofDatasetEphy The sliced object.
- inst
Examples
>>> # define the dataset >>> ds = DatasetEphy(...) >>> # temporal slicing of the data between (-100ms, 1500ms) >>> ds.sel(times=slice(-0.1, 1.5))
Examples using
sel:
- property times#
Time vector.
- property x#
Multi-subjects electrophysiological data (DataArray).
Examples using frites.dataset.DatasetEphy#
Define an electrophysiological dataset using MNE-Python structures
Define an electrophysiological dataset using Xarray
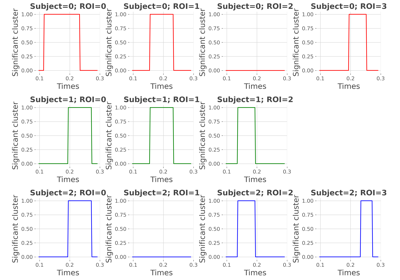
Compute a conjunction analysis on mutual-information
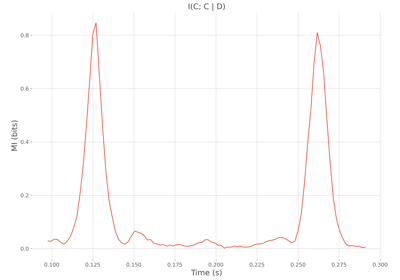
MI between two continuous variables conditioned by a discret one
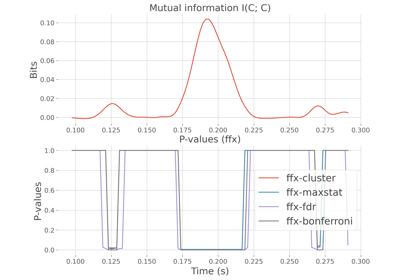
Compare within-subjects statistics when computing mutual information
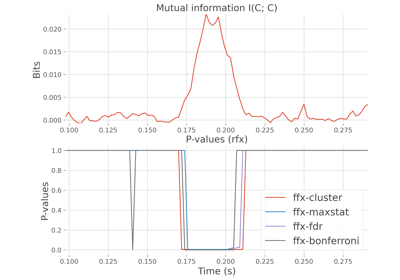
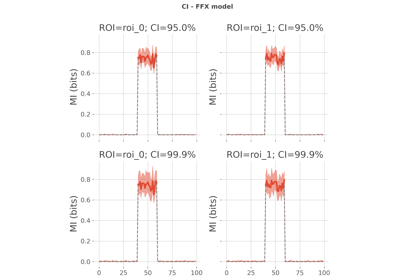
Compare between-subjects statistics when computing mutual information
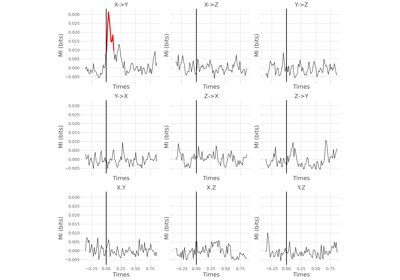
Statistical analysis of a stimulus-specific network