frites.conn.conn_dfc#
- frites.conn.conn_dfc(data, win_sample=None, times=None, roi=None, agg_ch=False, estimator=None, gcrn=False, n_jobs=1, verbose=None, **kw_links)[source]#
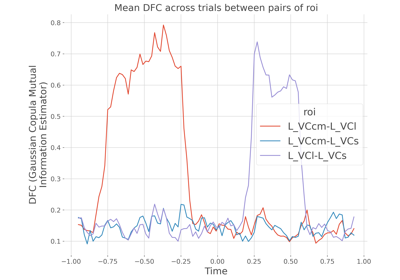
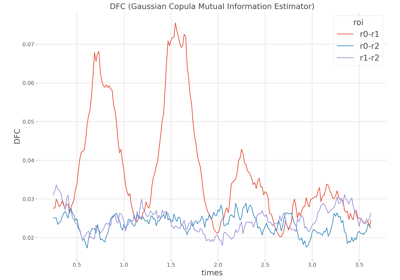
Single trial Dynamic Functional Connectivity.
This function computes the pairwise Dynamic Functional Connectivity (DFC) by estimating the statistical dependencies between time-series (possibly on sliding windows) and at the single-trial level using a measure of information. By default, if no estimator are provided the information shared between the time-series of two brain regions is estimated using the Gaussian-Copula Mutual Information (GCMI). However, other estimators can be provided (e.g correlation, distance correlation etc.)
- Parameters:
- datanumpy:array_like
Electrophysiological data. Several input types are supported :
Standard NumPy arrays of shape (n_epochs, n_roi, n_times)
mne.Epochs
xarray.DataArray of shape (n_epochs, n_roi, n_times)
- win_samplenumpy:array_like |
python:None Array of shape (n_windows, 2) describing where each window start and finish. You can use the function
frites.conn.define_windows()to define either manually either sliding windows. If None, the entire time window is used instead.- timesnumpy:array_like |
python:None Time vector array of shape (n_times,). If the input is an xarray, the name of the time dimension can be provided
- roinumpy:array_like |
python:None ROI names of a single subject. If the input is an xarray, the name of the ROI dimension can be provided
- agg_chbool |
python:False In case there are multiple electrodes, channels, contacts or sources inside a brain region, specify how the data has to be aggregated. Use either :
agg_ch=False : compute the pairwise DFC aross all possible pairs
agg_ch=True : compute the multivariate information
Note that feature is only available for measures of information supporting multivariate computations.
- estimator
frites.estimator|python:None Estimator in order to measure the amount of information shared between two time-series coming from two distinct brain regions. Note that if you want to privide an estimator, be sure that it is made for continuous variables (mi_type=’cc’). By default the Gaussian-Copula mutual-information is used.
- n_jobs
python:int| 1 Number of jobs to use for parallel computing (use -1 to use all jobs). The parallel loop is set at the pair level.
- kw_links
python:dict| {} Additional arguments for selecting links to compute are passed to the function
frites.conn.conn_links()
- Returns:
- dfcnumpy:array_like
The DFC array of shape (n_epochs, n_pairs, n_windows)
See also
Examples using frites.conn.conn_dfc#
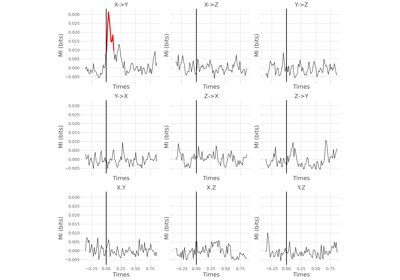
Statistical analysis of a stimulus-specific network