Note
Go to the end to download the full example code.
Estimate dynamic functional connectivity#
This example illustrates how to compute the dynamic functional connectivity (DFC) using the mutual information (MI). This type of connectivity is computed for each trial either inside a single window or across multiple windows.
import numpy as np
from itertools import product
from frites.simulations import sim_single_suj_ephy
from frites.conn import conn_dfc, define_windows, plot_windows
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Simulate electrophysiological data#
Let’s start by simulating MEG / EEG electrophysiological data coming from a single subject. The output data of this single subject has a shape of (n_epochs, n_roi, n_times)
modality = 'meeg'
n_roi = 3
n_epochs = 50
n_times = 1000
x, roi, _ = sim_single_suj_ephy(n_epochs=n_epochs, n_times=n_times,
modality=modality, n_roi=n_roi, random_state=0)
times = np.linspace(-1, 1, n_times)
Simulate spatial correlations#
Bellow, we are simulating some distant correlations by injecting the activity of an ROI to another
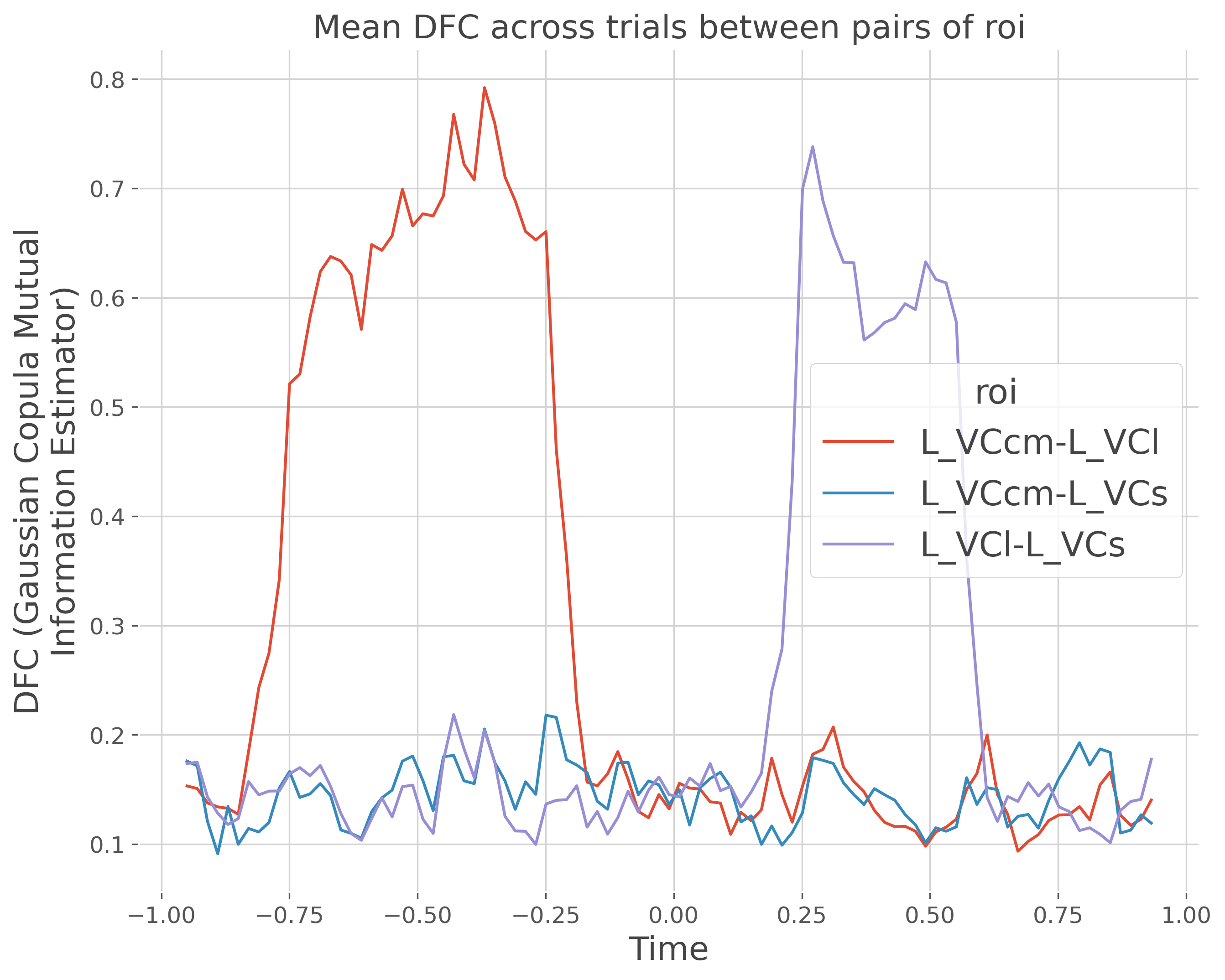
x[:, [1], slice(100, 400)] += x[:, [0], slice(100, 400)]
x[:, [2], slice(600, 800)] += x[:, [1], slice(600, 800)]
print(f'Corr 1 : {roi[0]}-{roi[1]} between [{times[100]}-{times[400]}]')
print(f'Corr 2 : {roi[2]}-{roi[1]} between [{times[600]}-{times[800]}]')
Corr 1 : L_VCcm-L_VCl between [-0.7997997997997999--0.19919919919919926]
Corr 2 : L_VCs-L_VCl between [0.20120120120120122-0.6016016016016015]
Define sliding windows#
Next, we define, and plot sliding windows in order to compute the DFC on consecutive time windows. In this example we used windows of length 100ms and 5ms between each consecutive windows
slwin_len = .1 # 100ms window length
slwin_step = .02 # 20ms between consecutive windows
win_sample = define_windows(times, slwin_len=slwin_len,
slwin_step=slwin_step)[0]
times_p = times[win_sample].mean(1)
# plt.figure(figsize=(10, 8))
# plot_windows(times, win_sample, title='Sliding windows')
# plt.ylim(-1, 1)
# plt.show()
Compute the DFC#
The DFC is going to be computed per trials, bewteen pairs of ROI and inside each of the temporal window
# compute DFC
dfc = conn_dfc(x, win_sample, times=times, roi=roi, n_jobs=1)
print(dfc)
plt.figure(figsize=(10, 8))
# plt.plot(times_p, dfc.mean('trials').T)
dfc.mean('trials').plot.line(x='times', hue='roi')
plt.xlabel('Time')
plt.title("Mean DFC across trials between pairs of roi")
plt.show()
0%| | Estimating DFC : 0/3 [00:00<?, ?it/s]
67%|██████▋ | Estimating DFC : 2/3 [00:00<00:00, 87.01it/s]
100%|██████████| Estimating DFC : 3/3 [00:00<00:00, 88.14it/s]
<xarray.DataArray 'DFC (Gaussian Copula Mutual Information Estimator)' (
trials: 50,
roi: 3,
times: 95)>
array([[[4.74126835e-04, 4.86970457e-05, 1.15753941e-01, ...,
1.44106135e-01, 1.00405946e-01, 8.37376192e-02],
[3.29334080e-01, 3.09351981e-01, 7.13511854e-02, ...,
6.81263686e-04, 2.51765386e-03, 3.19818184e-02],
[9.04738065e-03, 2.04119571e-02, 2.55241338e-02, ...,
6.54823380e-03, 1.80942327e-01, 7.69555792e-02]],
[[2.43588135e-01, 4.48731631e-02, 3.06968745e-02, ...,
1.19652078e-02, 1.41519258e-05, 1.56958587e-02],
[1.19740553e-01, 4.07272950e-02, 1.60232615e-02, ...,
4.46936190e-02, 5.86355664e-02, 7.60393683e-04],
[1.73070922e-01, 1.90565914e-01, 3.83086987e-02, ...,
1.16546720e-01, 1.21132061e-01, 3.26294273e-01]],
[[3.59413236e-01, 6.76742554e-01, 8.00896406e-01, ...,
2.62994856e-01, 3.78555506e-01, 3.44151706e-01],
[6.52924832e-03, 1.69114739e-01, 2.32962683e-01, ...,
2.62315989e-01, 3.79843652e-01, 1.67509735e-01],
[8.95062694e-04, 4.41071056e-02, 2.38820821e-01, ...,
2.81423610e-02, 7.95110539e-02, 4.08294201e-01]],
...
[[2.07532123e-02, 3.34448464e-06, 3.19930399e-03, ...,
2.95763791e-01, 1.60798971e-02, 1.35861740e-01],
[6.86131716e-01, 4.58753556e-01, 1.49167523e-01, ...,
3.07976492e-02, 1.27379432e-01, 6.57524914e-02],
[1.18787738e-03, 4.60020150e-04, 3.37673649e-02, ...,
2.37936508e-02, 1.87671706e-01, 1.34078279e-01]],
[[7.02991262e-02, 7.39582926e-02, 3.76868337e-01, ...,
1.00256223e-02, 2.14382401e-03, 1.74317230e-02],
[1.44385636e-01, 1.77252650e-01, 2.52936292e-03, ...,
2.17671096e-02, 1.07206358e-02, 2.39495244e-02],
[8.77984054e-03, 1.95415434e-03, 1.42009801e-03, ...,
3.53378356e-01, 3.31914216e-01, 4.86238658e-01]],
[[6.26457098e-04, 6.44705370e-02, 4.74612694e-03, ...,
5.78350900e-03, 7.82634515e-07, 6.35968670e-02],
[6.88552320e-01, 6.04924738e-01, 5.80820143e-02, ...,
7.78488629e-03, 6.61413297e-02, 2.66898096e-01],
[4.23928313e-02, 8.31796043e-03, 1.45379873e-02, ...,
1.08158283e-01, 9.71827563e-03, 2.18945459e-01]]])
Coordinates:
* trials (trials) int64 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* roi (roi) <U12 'L_VCcm-L_VCl' 'L_VCcm-L_VCs' 'L_VCl-L_VCs'
* times (times) float64 -0.9499 -0.9299 -0.9099 ... 0.8919 0.9119 0.9319
Attributes:
win_sample: [ 0 50 10 60 20 70 30 80 40 90 50 100 60 110 70...
win_times: [-0.94994995 -0.92992993 -0.90990991 -0.88988989 -0.86986987...
agg_ch: 0
type: dfc
estimator: Gaussian Copula Mutual Information Estimator
unit: Bits
sfreq: 499.50000000001125
sources: [0, 0, 1]
targets: [1, 2, 2]
Total running time of the script: (0 minutes 1.967 seconds)
Estimated memory usage: 395 MB