Note
Go to the end to download the full example code.
Estimate interaction information#
This example illustrates how to compute the interaction information (II) to investigate if pairs of brain regions are mainly carrying redundant or synergistic information about task-related variables (e.g. stimulus type, outcomes, learning rate etc.)
import numpy as np
import xarray as xr
from frites.simulations import sim_single_suj_ephy
from frites.conn import conn_ii
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Introduce redundant and synergistic informations#
No we can inject in the simulated data redundancy and synergy. We first start by defining a task-related variable (e.g. the learning rate) and we will then inject this variable into the two first brain regions to simulate redundant coding. Then, to simulate synergy, we will inject half of the trials of the learning rate into one brain region and the other half of trials to a different brain region. That way, the two brain regions are going to be needed to fully decode the learning rate which is going to be reflected by synergistic interactions.
# simulate the brain data
n_roi = 4
n_epochs = 200
n_times = 1000
x = np.random.rand(n_epochs, n_roi, n_times)
# let's start by defining the task-related variable
y = np.random.rand(n_epochs)
# reshape it to inject it inside the different brain regions
y_repeated = np.tile(y.reshape(-1, 1), (1, 100))
y_repeated *= np.hanning(y_repeated.shape[-1]).reshape(1, -1)
# inject y inside the first and second brain regions to introduce redundancy
# between them
t_half = int(np.round(n_times / 2))
x[:, 0, t_half - 50:t_half + 50] += y_repeated
x[:, 1, t_half - 50:t_half + 50] += y_repeated
# now, inject half of the trials inside the third brain region and the second
# half of the trils in the fourth region. That way, we'll introduce synergy
# between them as the two brain regions are going o be needed to fully decode
# y
x[0:50:, 2, t_half - 50:t_half + 50] += y_repeated[0:50, ...]
x[50::, 3, t_half - 50:t_half + 50] += y_repeated[50::, ...]
# finally, merge everything inside a data array
times = (np.arange(n_times) - t_half + 50) / 512.
roi = ['roi_1', 'roi_2', 'roi_3', 'roi_4']
x = xr.DataArray(x, dims=('trials', 'roi', 'times'), coords=(y, roi, times))
Compute the interaction information#
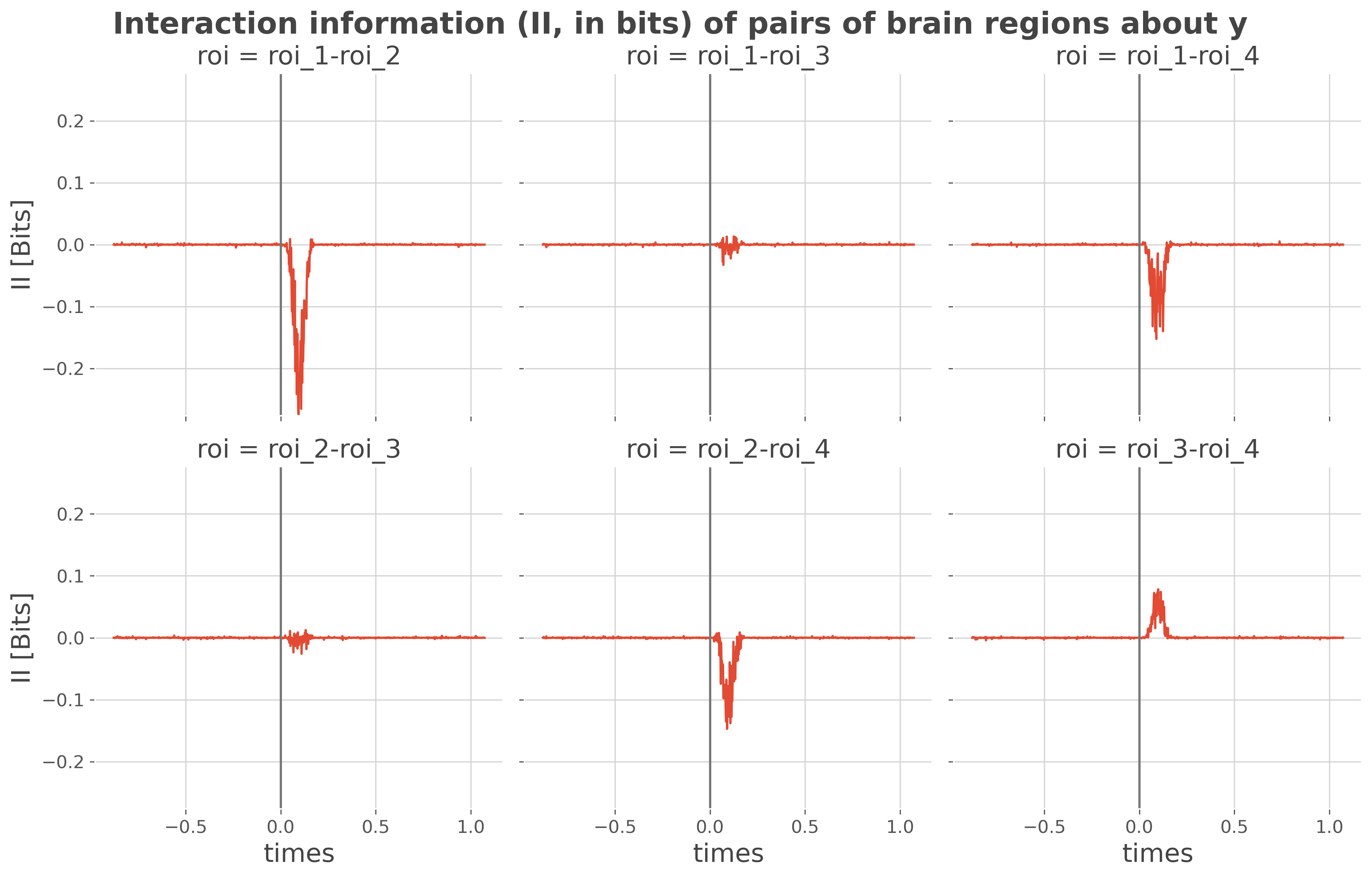
Now, we can estimate the interaction information. The II reflects whether pairs of brain regions are mainly carrying the same (i.e. redundant) information about the behavior or, if they are carrying complementary (i.e. synergy) information. Synergy is represented by a positive II and redundancy by a negative II.
# compute the II
ii = conn_ii(
x, 'trials', roi='roi', times='times', mi_type='cc'
)
print(ii)
# plot the result
fg = ii.plot(x='times', col='roi', col_wrap=3, size=4)
minmax = max(abs(ii.data.min()), abs(ii.data.max()))
plt.ylim(-minmax, minmax)
_ = [ax.axvline(0., color='C3') for ax in np.ravel(fg.axs)]
plt.gcf().suptitle(
"Interaction information (II, in bits) of pairs of brain regions about y",
fontsize=20, fontweight='bold', y=1.02)
plt.show()
0%| | Estimating MI on each node I(X;S) : 0/10 [00:00<?, ?it/s]
50%|█████ | Estimating total information I(X,Y;S) : 5/10 [00:00<00:00, 304.16it/s]
100%|██████████| Estimating total information I(X,Y;S) : 10/10 [00:00<00:00, 302.36it/s]
100%|██████████| Estimating total information I(X,Y;S) : 10/10 [00:00<00:00, 293.35it/s]
<xarray.DataArray 'II' (roi: 6, times: 1000)>
array([[-3.55877346e-05, 1.07177756e-03, 3.57478593e-05, ...,
-2.06468240e-05, 1.75047855e-05, -9.71695737e-06],
[ 5.19280277e-06, 4.79825541e-05, 1.17727070e-03, ...,
-1.98780782e-05, -1.41096471e-04, -3.73062125e-05],
[-2.86675497e-05, 7.98898509e-04, -3.58633755e-04, ...,
-1.76841740e-06, 3.21195583e-04, -1.50849164e-05],
[ 7.62301673e-05, -7.39087452e-06, -4.78554388e-04, ...,
1.47675307e-04, -7.73725523e-05, -1.51109463e-04],
[ 1.39735429e-04, 6.37082269e-04, 1.31262167e-03, ...,
-1.13274486e-04, -1.34005129e-04, -2.09395359e-05],
[-7.71568707e-05, -1.21715388e-05, 1.28502118e-03, ...,
-6.42886884e-04, -4.54571259e-04, 1.70420687e-04]])
Coordinates:
* roi (roi) <U11 'roi_1-roi_2' 'roi_1-roi_3' ... 'roi_3-roi_4'
* times (times) float64 -0.8789 -0.877 -0.875 -0.873 ... 1.068 1.07 1.072
Attributes:
sfreq: 512.0
sources: [0, 0, 0, 1, 1, 2]
targets: [1, 2, 3, 2, 3, 3]
mi_type: cc
gcrn: 1
dt: 1
unit: Bits
As we see in the results : - The II between roi_1 and roi_2 is redundant (II < 0) - The II between roi_3 and roi_4 is synergistic (II > 0) - The remaining pairs of brain regions are mainly carrying slightly redundant
information
Total running time of the script: (0 minutes 2.058 seconds)
Estimated memory usage: 395 MB