Note
Go to the end to download the full example code.
MI between two continuous variables#
This example illustrates how to compute the mutual information between two continuous variables and also perform statistics. Usually, the first variable is an electrophysiological data (M/EEG, intracranial) and a regressor. This kind of mutual information is equivalent to a traditional correlation. Note that the regressor variable can either be univariate (single column) or multivariate (multiple columns). For further details, see Ince et al., 2017 [12]
import numpy as np
from frites.simulations import sim_multi_suj_ephy, sim_mi_cc
from frites.dataset import DatasetEphy
from frites.workflow import WfMi
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Simulate electrophysiological data#
Let’s start by simulating MEG / EEG electrophysiological data coming from
multiple subjects using the function
frites.simulations.sim_multi_suj_ephy(). As a result, the x output
is a list of length n_subjects of arrays, each one with a shape of
n_epochs, n_sites, n_times
modality = 'meeg'
n_subjects = 5
n_epochs = 400
n_times = 100
x, roi, time = sim_multi_suj_ephy(n_subjects=n_subjects, n_epochs=n_epochs,
n_times=n_times, modality=modality,
random_state=0)
Extract the regressor variable#
Once we have the electrophysiological, we need to extract the second variable that is going to serves for computing the “correlation” at each time point and at each site / channel / sensor. To do this, we can simply take the mean over time points and region of interest in a time window
sl = slice(40, 60)
y = [x[k][..., sl].mean(axis=(1, 2)) for k in range(len(x))]
Note
Taking the mean across time points and space is exactly the behavior of
the function frites.simulations.sim_mi_cc()
Define the electrophysiological dataset#
Now we define an instance of frites.dataset.DatasetEphy
dt = DatasetEphy(x.copy(), y=y, roi=roi, times=time)
Compute the mutual information#
Once we have the dataset instance, we can then define an instance of workflow
frites.workflow.WfMi. This instance is used to compute the mutual
information
# mutual information type ('cc' = continuous / continuous)
mi_type = 'cc'
# define the workflow
wf = WfMi(mi_type, inference='ffx')
# compute the mutual information without permutations
mi, _ = wf.fit(dt, mcp=None)
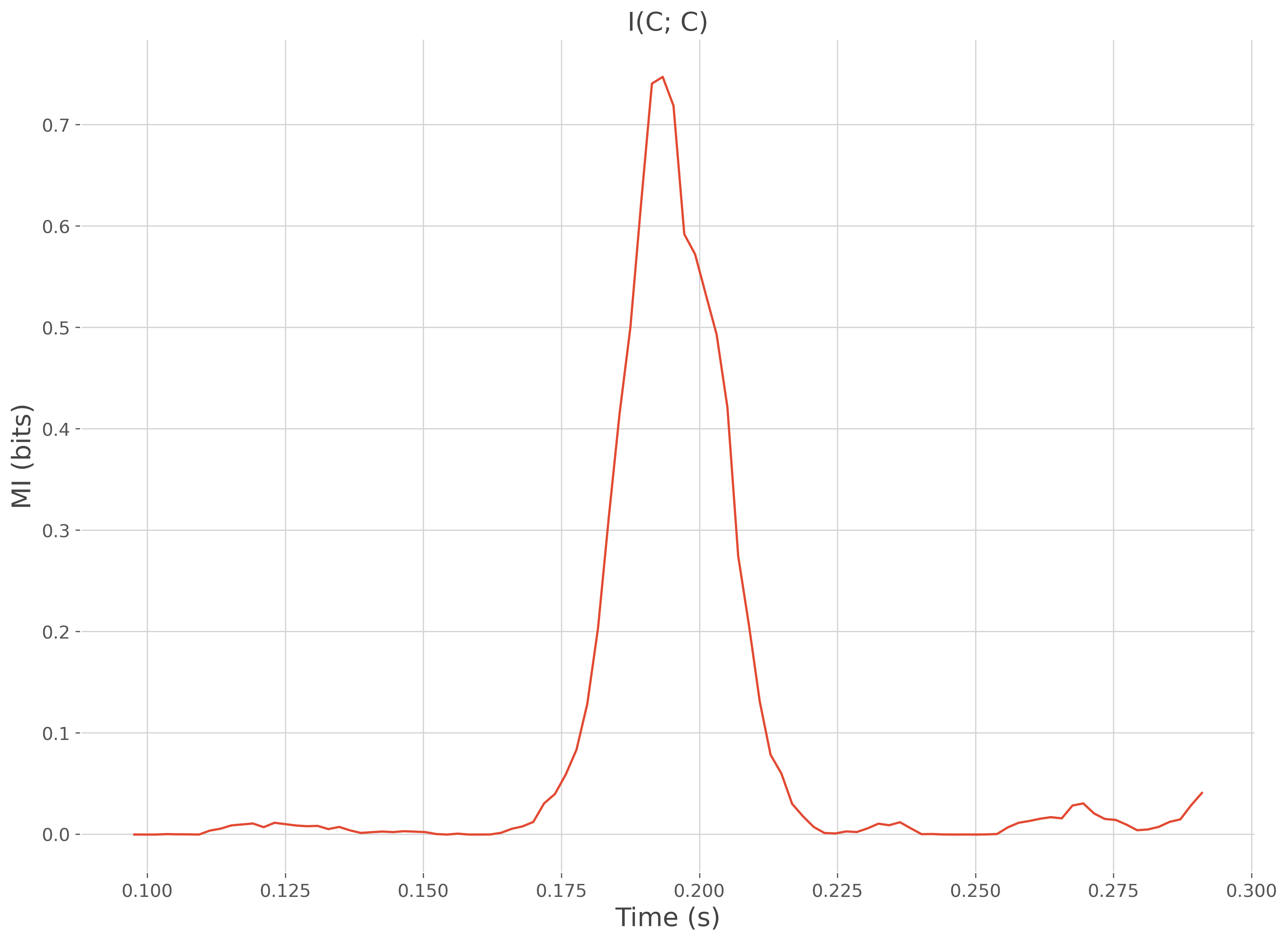
# plot the information shared between the data and the regressor y
plt.plot(time, mi)
plt.xlabel("Time (s)"), plt.ylabel("MI (bits)")
plt.title('I(C; C)')
plt.show()
0%| | Estimating MI : 0/1 [00:00<?, ?it/s]
100%|██████████| Estimating MI : 1/1 [00:00<00:00, 13.79it/s]
100%|██████████| Estimating MI : 1/1 [00:00<00:00, 13.66it/s]
Evaluate the statistics#
In the section above, the input parameter stat_method=None specifies that no statistics are going to be computed. Here, we show how to compute either within (ffx) or between subject (rfx) statistics.
mi_type = 'cc'
n_perm = 200
y, _ = sim_mi_cc(x, snr=.1)
# within subject statistics (ffx=fixed-effect)
ffx_stat = 'ffx_cluster_tfce'
dt_ffx = DatasetEphy(x.copy(), y=y, roi=roi, times=time)
wf_ffx = WfMi(mi_type=mi_type, inference='ffx')
mi_ffx, pv_ffx = wf_ffx.fit(dt_ffx, mcp='cluster', cluster_th='tfce',
n_perm=n_perm, n_jobs=1)
# between-subject statistics (rfx=random-effect)
dt_rfx = DatasetEphy(x.copy(), y=y, roi=roi, times=time)
wf_rfx = WfMi(mi_type=mi_type, inference='rfx')
mi_rfx, pv_rfx = wf_rfx.fit(dt_rfx, mcp='cluster', cluster_th='tfce',
n_perm=n_perm, n_jobs=1)
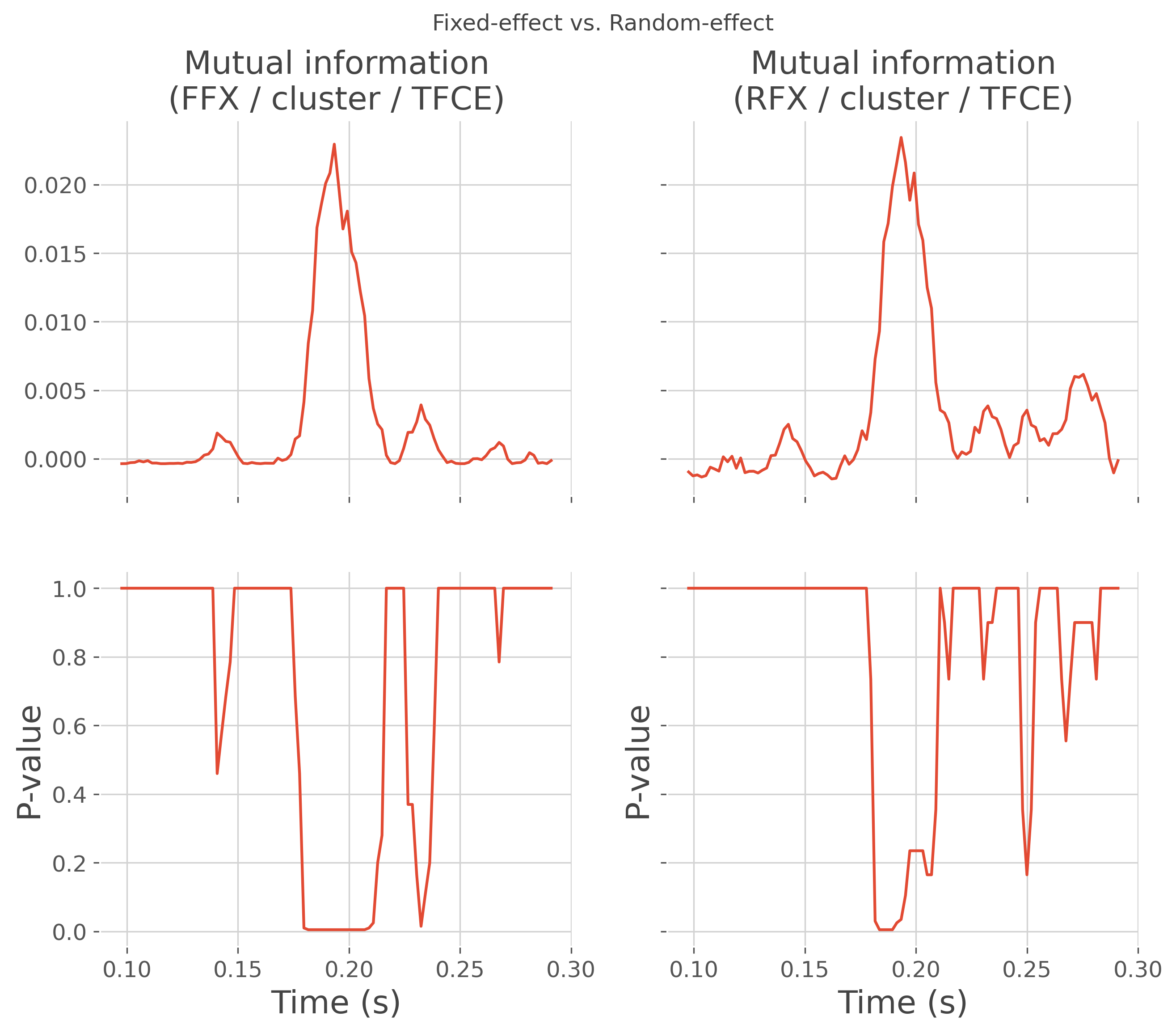
# plot the comparison
fig, axs = plt.subplots(nrows=2, ncols=2, sharex=True, sharey='row',
figsize=(10, 8))
fig.suptitle("Fixed-effect vs. Random-effect")
plt.sca(axs[0, 0])
plt.plot(time, mi_ffx)
plt.title(f"Mutual information\n(FFX / cluster / TFCE)")
plt.sca(axs[1, 0])
plt.plot(time, pv_ffx)
plt.xlabel("Time (s)"), plt.ylabel("P-value")
plt.sca(axs[0, 1])
plt.plot(time, mi_rfx)
plt.title(f"Mutual information\n(RFX / cluster / TFCE)")
plt.sca(axs[1, 1])
plt.plot(time, pv_rfx)
plt.xlabel("Time (s)"), plt.ylabel("P-value")
plt.show()
0%| | Estimating MI : 0/1 [00:00<?, ?it/s]
100%|██████████| Estimating MI : 1/1 [00:00<00:00, 3.43it/s]
100%|██████████| Estimating MI : 1/1 [00:00<00:00, 3.43it/s]
0%| | Estimating MI : 0/1 [00:00<?, ?it/s]
100%|██████████| Estimating MI : 1/1 [00:01<00:00, 1.59s/it]
100%|██████████| Estimating MI : 1/1 [00:01<00:00, 1.59s/it]
Total running time of the script: (0 minutes 5.723 seconds)
Estimated memory usage: 395 MB