Note
Go to the end to download the full example code.
FIT: Feature specific information transfer#
This example illustrates how to compute Feature-specific Information
Transfer (FIT), quantifying how much information about a specific
feature flows between two regions. FIT merges the Wiener-Granger causality
principle with information-content specificity.
The theoretical background is described in [1] and the FIT is computed
using the frites.conn.conn_fit() function.
[1] Celotto M, Bím J, Tlaie A, De Feo V, Toso A, Lemke SM, Chicharro D,
Nili H, Bieler M, Hanganu-Opatz IL, Donner TH, Brovelli A, Panzeri S
(2023). An information-theoretic quantification of the content of
communication between brain regions. Advances in Neural Information
Processing Systems (NeurIPS), 36, 64213-64265
import numpy as np
import xarray as xr
from frites.simulations import StimSpecAR
from frites.conn import conn_fit
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Data simulation#
Here, we use an auto-regressive simulating a gamma increase.
net = False
avg_delay = False
ar_type = 'hga'
n_stim = 3
n_epochs = 400
ss = StimSpecAR()
ar = ss.fit(ar_type=ar_type, n_epochs=n_epochs, n_stim=n_stim, random_state=0)
print(ar)
plt.figure(figsize=(7, 8))
ss.plot(cmap='bwr')
plt.tight_layout()
plt.show()
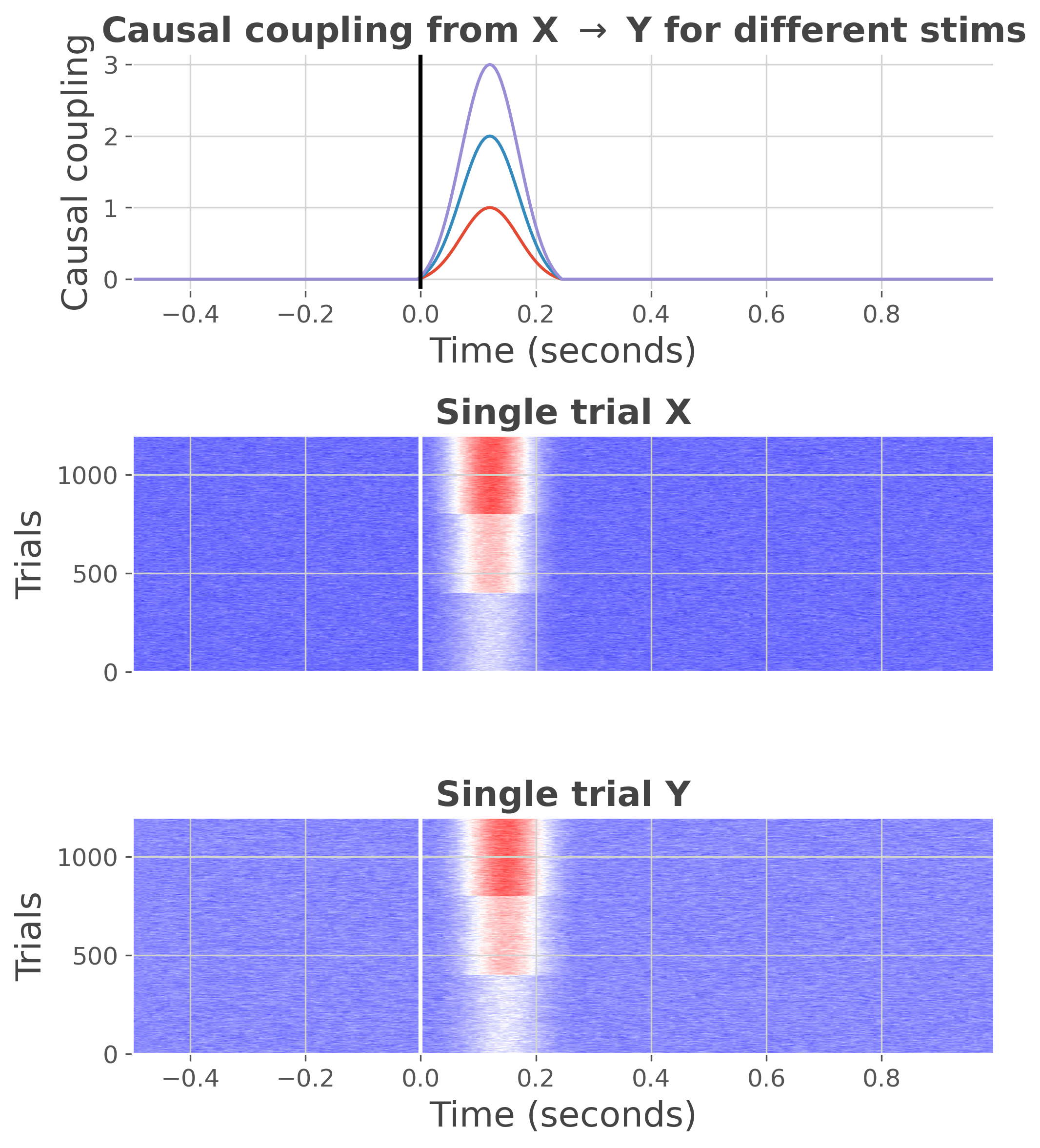
<xarray.DataArray (trials: 1200, roi: 2, times: 300)>
array([[[ 0.3944541 , 0.08947787, 0.21885247, ..., 0.15821295,
-0.04185563, 0.07019032],
[ 0.36321467, -0.13679289, -0.11810279, ..., 0.36246378,
-0.15565568, 0.14413607]],
[[-0.29214829, 0.37076929, -0.02642228, ..., -0.07226816,
-0.22966976, -0.10883466],
[ 0.46192319, -0.32896069, -0.18563208, ..., 0.1989966 ,
-0.12743325, 0.30140249]],
[[-0.34668654, 0.09331533, -0.21116721, ..., -0.37032276,
0.46903415, 0.05036588],
[-0.23992211, 0.11072083, -0.21288756, ..., -0.28649571,
-0.07005528, -0.34033346]],
...,
[[-0.01538188, -0.16898504, 0.23711941, ..., -0.01158757,
-0.12814105, -0.06427391],
[ 0.3718958 , 0.09224002, -0.202277 , ..., -0.05967159,
-0.47515636, 0.06669148]],
[[ 0.2289687 , 0.26491207, -0.28432468, ..., 0.44090788,
0.19124113, 0.14904106],
[ 0.18744301, -0.09694941, 0.39603464, ..., 0.05478838,
0.18196749, -0.0345686 ]],
[[-0.06437222, -0.23064244, -0.08996951, ..., 0.0400626 ,
0.0977585 , -0.12655847],
[ 0.04681595, 0.05029251, 0.1605935 , ..., 0.1253402 ,
0.14925333, -0.22551455]]])
Coordinates:
* trials (trials) int64 1 1 1 1 1 1 1 1 1 1 1 1 ... 3 3 3 3 3 3 3 3 3 3 3 3
* roi (roi) <U1 'x' 'y'
* times (times) float64 -0.5 -0.495 -0.49 -0.485 ... 0.98 0.985 0.99 0.995
Attributes:
n_stim: 3
n_std: 3
ar_type: hga
stimulus: [1 2 3]
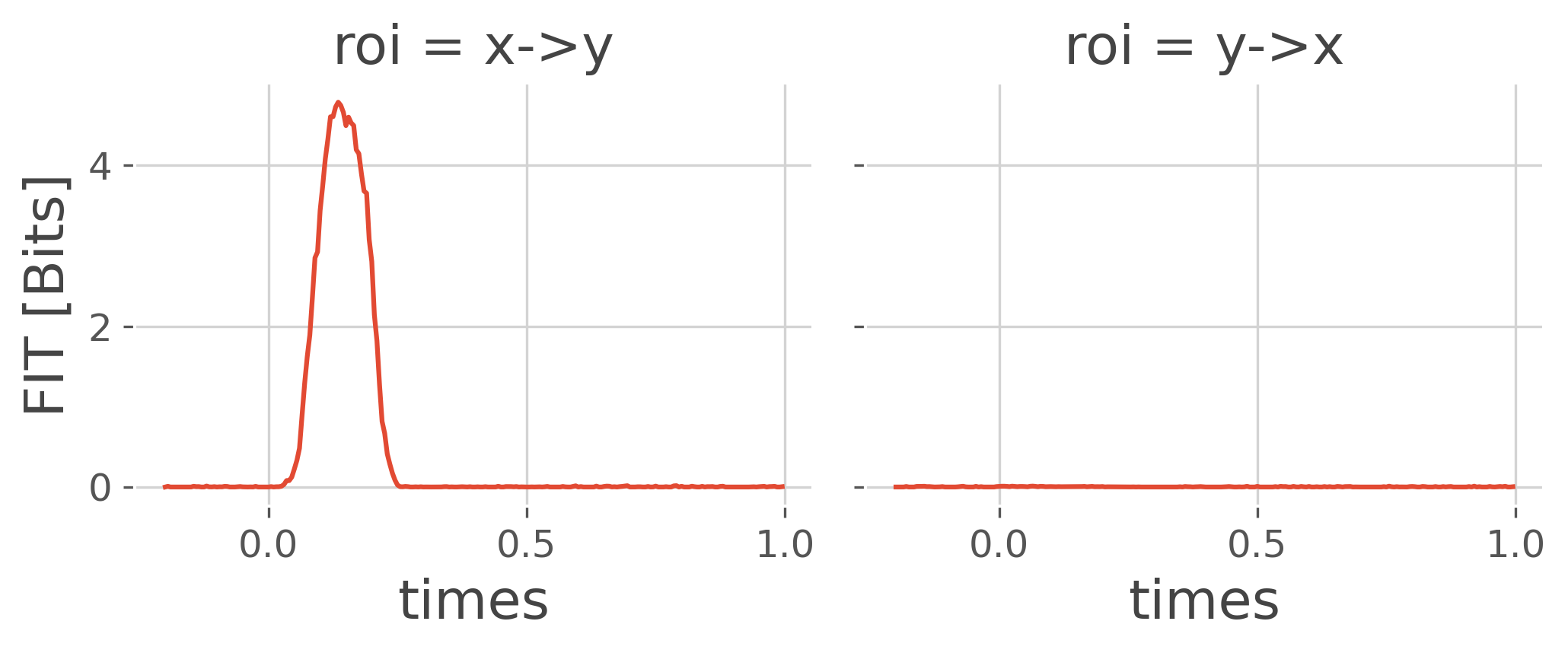
Compute Feature specific information transfer#
Now we can use the simulated data to estimate the FIT.
# Compute the FIT
fit = conn_fit(ar, y='trials', roi='roi', times='times', mi_type='cd',
max_delay=.3, net=net, verbose=False, avg_delay=avg_delay)
# Plot the results
fit.plot(x='times', col='roi') # net = False
plt.show()
Total running time of the script: (0 minutes 2.815 seconds)
Estimated memory usage: 456 MB