Note
Go to the end to download the full example code.
AR : simulate common driving input#
This example illustrates an autoregressive model that simulates a common driving input (i.e X->Y and X->Z) and how it is measured using the covariance based Granger Causality
import numpy as np
from frites.simulations import StimSpecAR
from frites.conn import conn_covgc
import matplotlib.pyplot as plt
Simulate 3 nodes 40hz oscillations#
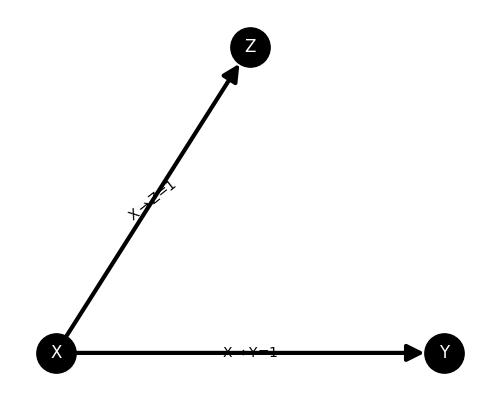
Here, we use the class frites.simulations.StimSpecAR to simulate an
stimulus-specific autoregressive model made of three nodes (X, Y and Z). This
network simulates a transfer X->Y and X->Z. X is then called a common driving
input for Y and Z
ar_type = 'osc_40_3' # 40hz oscillations
n_stim = 3 # number of stimulus
n_epochs = 50 # number of epochs per stimulus
ss = StimSpecAR()
ar = ss.fit(ar_type=ar_type, n_epochs=n_epochs, n_stim=n_stim)
print(ar)
<xarray.DataArray (trials: 150, roi: 3, times: 300)>
array([[[ 0.25134439, -0.04588286, -0.38997855, ..., 0.47525239,
-0.42996041, -0.35923828],
[ 0.28807864, 0.32943226, -0.15219402, ..., 0.13200512,
0.01481161, 0.18764986],
[ 0.23625422, 0.16773289, 0.12441169, ..., 0.21856348,
-0.46722405, -0.21981999]],
[[-0.23425387, 0.2383734 , 0.12571813, ..., 0.01028065,
0.86762484, 0.64894575],
[-0.00829905, -0.39008386, 0.08640293, ..., -0.17602529,
-0.10660233, -0.25474297],
[ 0.41032334, 0.10277509, -0.36499451, ..., -0.20049947,
-0.44564005, -0.14311654]],
[[-0.4885944 , -0.24088023, 0.5716417 , ..., 0.42614904,
0.27062738, -0.17958764],
[-0.35214581, -0.18335921, -0.04724976, ..., -0.00556085,
-0.25871338, -0.35545094],
[ 0.05442133, -0.08335093, 0.08436663, ..., -0.30889446,
-0.3294228 , 0.36430815]],
...
[[ 0.33821833, 0.13736941, -0.04772936, ..., -0.12733071,
-0.50344985, -0.06069676],
[-0.35159565, -0.06081666, 0.41416986, ..., 0.43957058,
-0.32255854, -0.14764446],
[-0.13518498, 0.10178025, 0.32153082, ..., 0.23252699,
-0.23642249, 0.07492754]],
[[-0.19209026, 0.07261479, 0.11361513, ..., -0.25965606,
-0.14892411, 0.10439557],
[ 0.43026152, -0.02417257, -0.32028906, ..., 0.12009966,
0.31882895, 0.14351507],
[-0.1940527 , -0.06980799, 0.18732115, ..., -0.37826542,
-0.36238163, 0.00259724]],
[[ 0.19648887, 0.05398445, -0.14429047, ..., -0.37445137,
-0.23278332, 0.33820427],
[-0.0631801 , -0.0741364 , -0.11359434, ..., 0.05873906,
-0.0372718 , 0.13327158],
[-0.10949838, 0.04815722, 0.04406834, ..., -0.03883177,
-0.31380632, -0.14499861]]])
Coordinates:
* trials (trials) int64 1 1 1 1 1 1 1 1 1 1 1 1 ... 3 3 3 3 3 3 3 3 3 3 3 3
* roi (roi) <U1 'x' 'y' 'z'
* times (times) float64 -0.5 -0.495 -0.49 -0.485 ... 0.98 0.985 0.99 0.995
Attributes:
n_stim: 3
n_std: 3
ar_type: osc_40_3
stimulus: [1 2 3]
plot the network
plt.figure(figsize=(5, 4))
ss.plot_model()
plt.show()
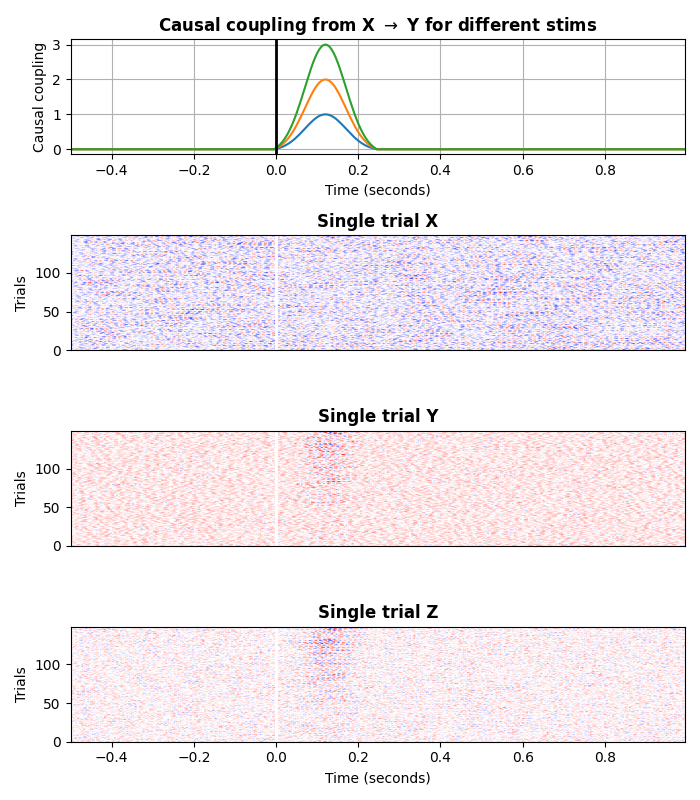
plot the data
plt.figure(figsize=(7, 8))
ss.plot(cmap='bwr')
plt.tight_layout()
plt.show()
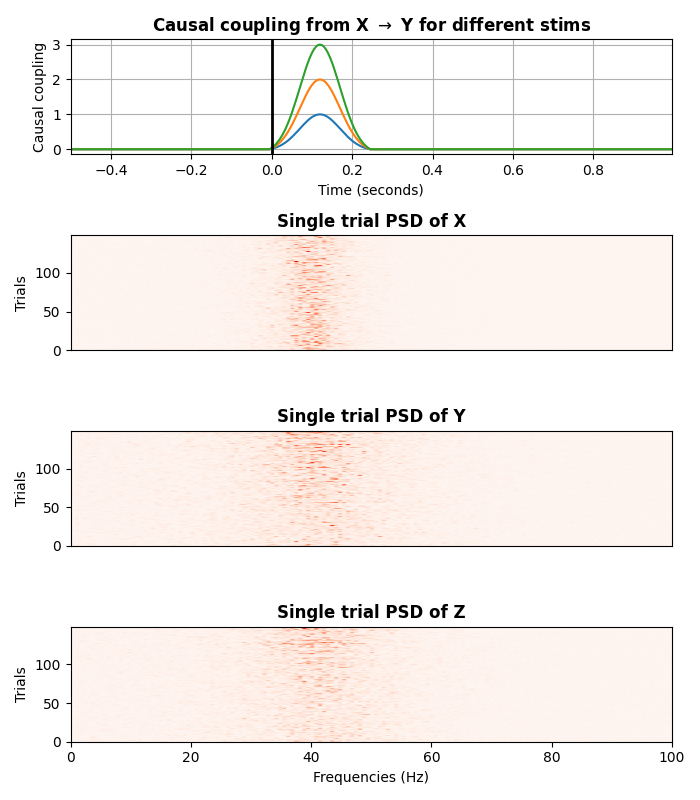
plot the power spectrum density (PSD)
plt.figure(figsize=(7, 8))
ss.plot(cmap='Reds', psd=True)
plt.tight_layout()
plt.show()
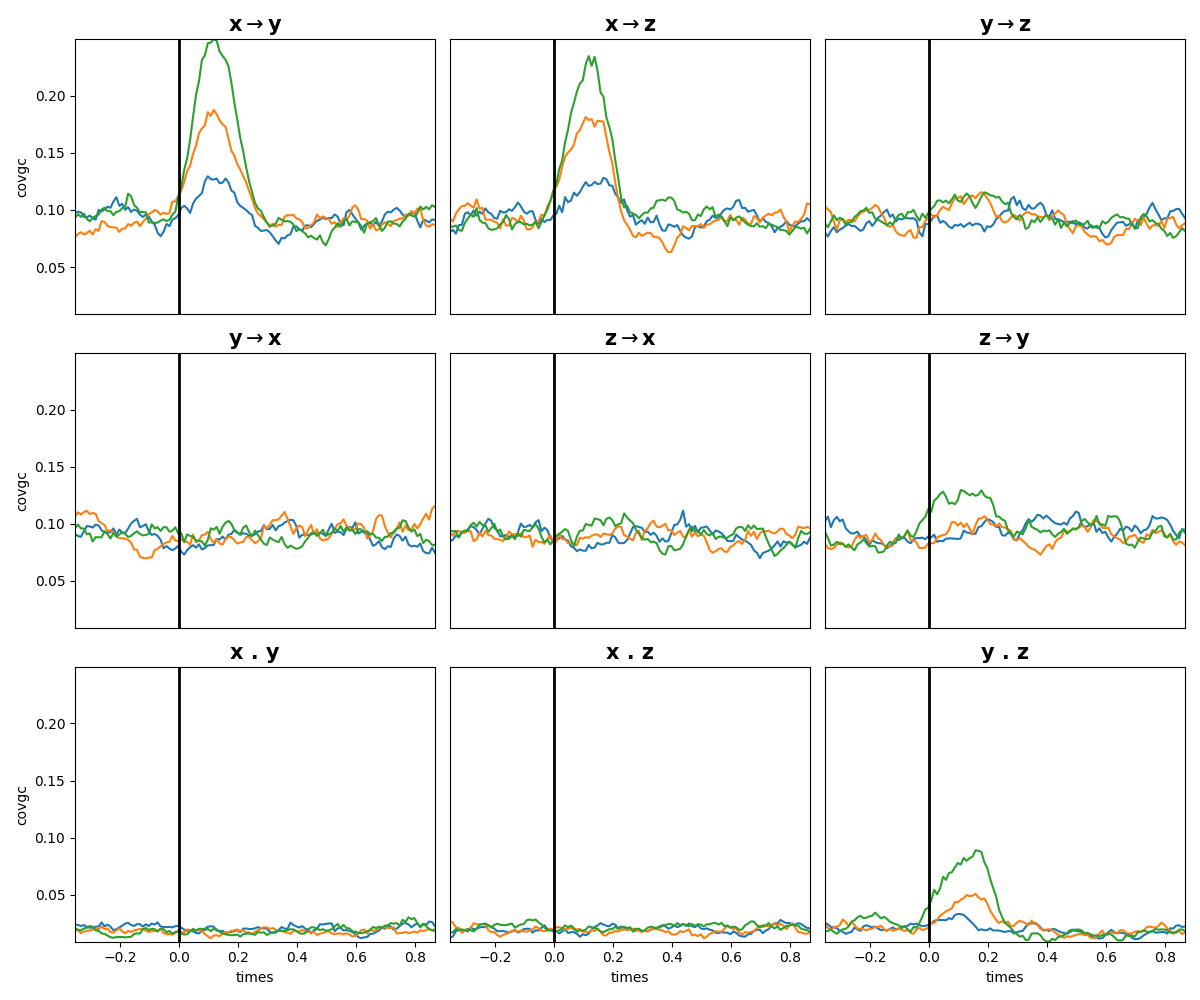
Compute the Granger-Causality#
We then compute and plot the Granger Causality. From the plot you can see that there’s indeed an information transfer from X->Y and X->Z and, in addition, an instantaneous connectivity between Y.Z
dt = 50
lag = 5
step = 2
t0 = np.arange(lag, ar.shape[-1] - dt, step)
gc = conn_covgc(ar, roi='roi', times='times', dt=dt, lag=lag, t0=t0,
n_jobs=-1)
# sphinx_gallery_thumbnail_number = 4
plt.figure(figsize=(12, 10))
ss.plot_covgc(gc)
plt.tight_layout()
plt.show()
0%| | : 0/3 [00:00<?, ?it/s]
100%|██████████| : 3/3 [00:00<00:00, 3376.15it/s]
Total running time of the script: (0 minutes 11.636 seconds)
Estimated memory usage: 401 MB