Note
Go to the end to download the full example code.
Compute a conjunction analysis on mutual-information#
In this example, we show how to perform a conjunction analysis which consist in getting the number of subjects that present significant effect.
import numpy as np
from frites.simulations import sim_single_suj_ephy
from frites.dataset import DatasetEphy
from frites.workflow import WfMi
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Define where to put an effect for each subject#
We start by organising where to set an affect for each subject. To this end, we define a dictionary where each key correspond to a subject. Then, for each subject we add the number of roi (n_roi), a color (color) and finally where to introduce effects ((roi_number, (effect_start, effect_end)))
n_epochs = 200
n_times = 100
ephy = {
'subject_0': {
'n_roi': 4,
'color': 'red',
'clusters': [(0, (10, 70)), (1, (30, 70)), (3, (50, 80))]
},
'subject_1': {
'n_roi': 3,
'color': 'green',
'clusters': [(0, (50, 90)), (1, (30, 70)), (2, (20, 50))]
},
'subject_2': {
'n_roi': 4,
'color': 'blue',
'clusters': [(0, (50, 90)), (2, (20, 50)), (3, (70, 90))]
}
}
n_subjects = len(ephy)
Generate the electrophysiological data#
Now we have all the properties where effect have to be present, we generate
the electrophysiological data using the function
frites.simulations.sim_single_suj_ephy().
# generate some random data
x, roi = [], []
for s_nb, s_name in enumerate(ephy.keys()):
n_roi = ephy[s_name]['n_roi']
_x, _roi, times = sim_single_suj_ephy(
modality="meeg", sf=512., n_times=n_times, n_roi=n_roi,
n_sites_per_roi=1, n_epochs=n_epochs, random_state=s_nb)
x += [_x]
roi += [np.array([f"roi_{k}" for k in range(n_roi)])]
# introduce an effect
y = []
for s_nb, s_name in enumerate(ephy.keys()):
_y = np.random.normal(size=(n_epochs, 1))
clusters = ephy[s_name]['clusters']
for roi_nb, t_idx in clusters:
x[s_nb][:, roi_nb, t_idx[0]:t_idx[1]] += _y
y += [_y.squeeze()]
# Define the electrophysiological dataset
dt = DatasetEphy(x, y=y, roi=roi, times=times)
Compute the mutual information#
Once we have the dataset instance, we can then define an instance of workflow
frites.workflow.WfMi. This instance is used to compute the mutual
information
# mutual information type ('cc' = continuous / continuous)
mi_type = 'cc'
inference = 'rfx' # don't use 'ffx' for assessing conjunction analysis !
# define the workflow
wf = WfMi(mi_type)
# compute the mutual information
mi, pv = wf.fit(dt, mcp='cluster', n_perm=200, n_jobs=1, random_state=0)
n_roi = len(mi.roi.data)
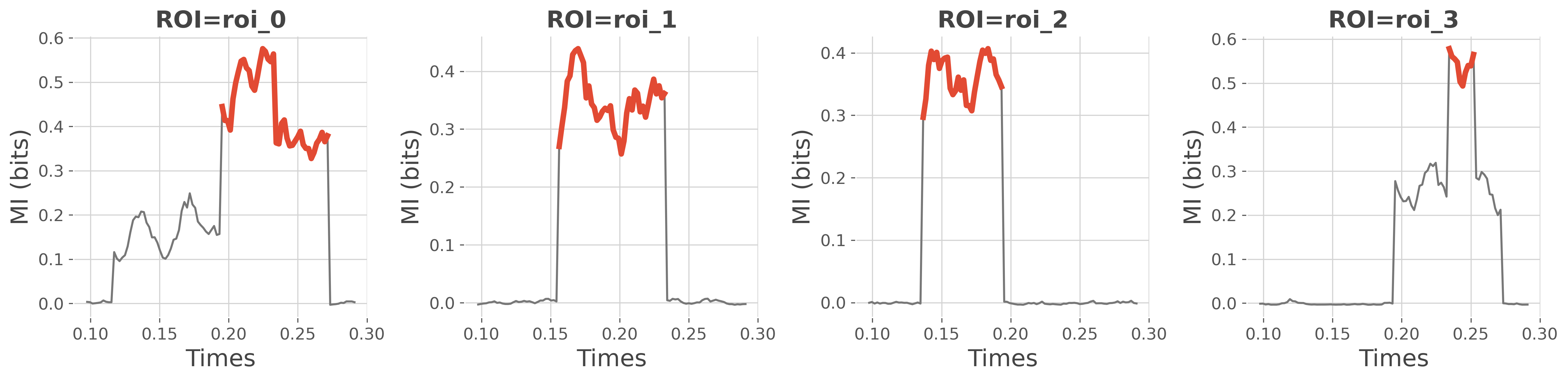
# plot where there's significant values of mi
fig = plt.figure(figsize=(16, 4))
for n_r, r in enumerate(mi.roi.data):
# select the mi and p-values a specific roi
mi_r, pv_r = mi.sel(roi=r), pv.sel(roi=r)
# make a copy of the mi and set to nan everywhere it's not significant
mi_sr = mi_r.copy()
mi_sr.data[pv_r >= .05] = np.nan
# superimpose mi and significant mi
plt.subplot(1, n_roi, n_r + 1)
plt.plot(times, mi_r, color='C3')
plt.plot(times, mi_sr, lw=4, color='C0')
plt.xlabel('Times'), plt.ylabel('MI (bits)')
plt.title(f"ROI={r}", fontweight='bold')
plt.tight_layout()
plt.show()
0%| | Estimating MI : 0/4 [00:00<?, ?it/s]
25%|██▌ | Estimating MI : 1/4 [00:00<00:01, 1.77it/s]
50%|█████ | Estimating MI : 2/4 [00:01<00:01, 1.78it/s]
75%|███████▌ | Estimating MI : 3/4 [00:01<00:00, 1.79it/s]
100%|██████████| Estimating MI : 4/4 [00:02<00:00, 1.95it/s]
100%|██████████| Estimating MI : 4/4 [00:02<00:00, 1.94it/s]
Perform the conjunction analysis#
Now we have the values of MI we can compute the conjunction analysis. The following method returns two DataArray :
- conj_ssDataArray of shape (n_subjects, n_times, n_roi) that contains the
significant MI of each subject
- conjDataArray of shape (n_times, n_roi) that contains the number of
subjects that have a significant effect at each time point and for each roi
# perform the conjunction analysis
conj_ss, conj = wf.conjunction_analysis()
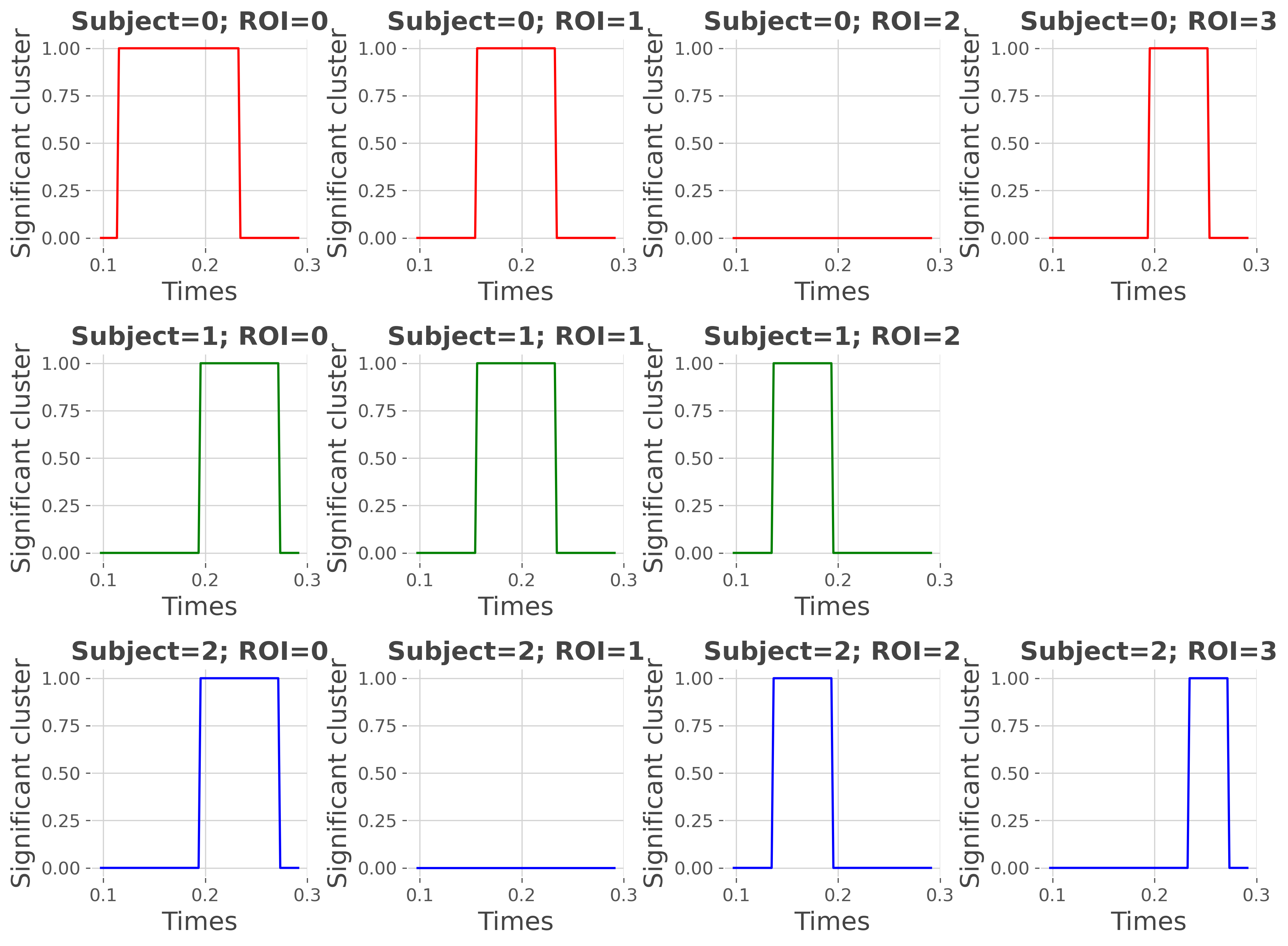
Plot where there’s significant effect for each subject#
# printing the results
print(conj_ss)
fig = plt.figure(figsize=(12, 9))
q = 0
for n_s in range(n_subjects):
color = ephy[f'subject_{n_s}']['color']
for n_r, roi in enumerate(conj.roi.data):
q += 1
ss_data = conj_ss.sel(roi=roi, subject=n_s).T
if np.isnan(ss_data).all():
continue
plt.subplot(n_subjects, n_roi, q)
plt.plot(times, ss_data, color=color)
plt.ylim(-0.05, 1.05)
plt.title(f"Subject={n_s}; ROI={n_r}", fontweight='bold')
plt.xlabel('Times'), plt.ylabel('Significant cluster')
plt.tight_layout()
plt.show()
<xarray.DataArray 'Single subject conjunction' (subject: 3, times: 100, roi: 4)>
array([[[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
...,
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.]],
[[ 0., 0., 0., nan],
[ 0., 0., 0., nan],
[ 0., 0., 0., nan],
...,
[ 0., 0., 0., nan],
[ 0., 0., 0., nan],
[ 0., 0., 0., nan]],
[[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
...,
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.],
[ 0., 0., 0., 0.]]])
Coordinates:
* times (times) float64 0.09766 0.09961 0.1016 ... 0.2891 0.291
* roi (roi) <U5 'roi_0' 'roi_1' 'roi_2' 'roi_3'
* subject (subject) int64 0 1 2
p float64 0.05
cluster_th <U4 'none'
cluster_alpha float64 0.05
mcp <U7 'cluster'
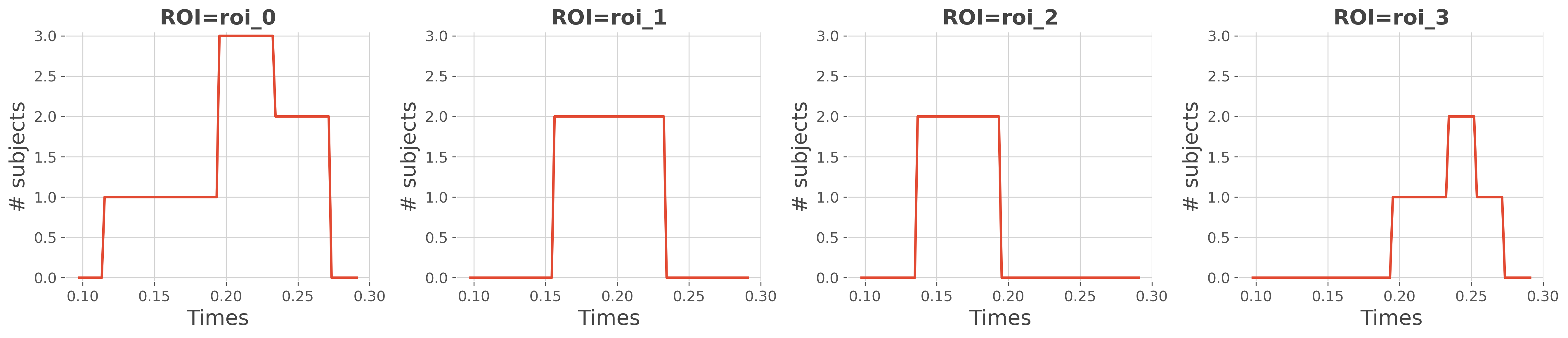
Plot the number of subjects that have an effect#
# printing the results
print(conj)
# sphinx_gallery_thumbnail_number = 2
fig = plt.figure(figsize=(18, 4))
for n_r, roi in enumerate(conj.roi.data):
plt.subplot(1, n_roi, n_r + 1)
plt.plot(times, conj.sel(roi=roi), lw=2)
plt.title(f"ROI={roi}", fontweight='bold')
plt.xlabel('Times'), plt.ylabel('# subjects')
plt.ylim(-0.05, n_subjects + 0.05)
plt.tight_layout()
plt.show()
<xarray.DataArray 'Across subjects conjunction' (times: 100, roi: 4)>
array([[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
...
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[2., 0., 0., 1.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.]])
Coordinates:
* times (times) float64 0.09766 0.09961 0.1016 ... 0.2891 0.291
* roi (roi) <U5 'roi_0' 'roi_1' 'roi_2' 'roi_3'
p float64 0.05
cluster_th <U4 'none'
cluster_alpha float64 0.05
mcp <U7 'cluster'
Total running time of the script: (0 minutes 8.200 seconds)
Estimated memory usage: 431 MB