Note
Go to the end to download the full example code.
AR : pairwise illustration#
This example illustrates a simple autoregressive model simulating a stimulus-specific information transfer from a source X to a target Y.
from frites import set_mpl_style
from frites.simulations import StimSpecAR
import matplotlib.pyplot as plt
set_mpl_style()
Simulate 40hz oscillations#
Here, we use the class frites.simulations.StimSpecAR to simulate an
stimulus-specific autoregressive model. For the pairwise models, you can
choose :
‘hga’ : high-gamma burst
‘osc_40’ / ‘osc_20’ : respectivelly 20hz and 40hz oscillations
‘ding_2’ : pairwise Ding’s model [7]
ar_type = 'osc_40' # 40hz oscillations
n_stim = 3 # number of stimulus
n_epochs = 50 # number of epochs per stimulus
ss = StimSpecAR()
ar = ss.fit(ar_type=ar_type, n_epochs=n_epochs, n_stim=n_stim)
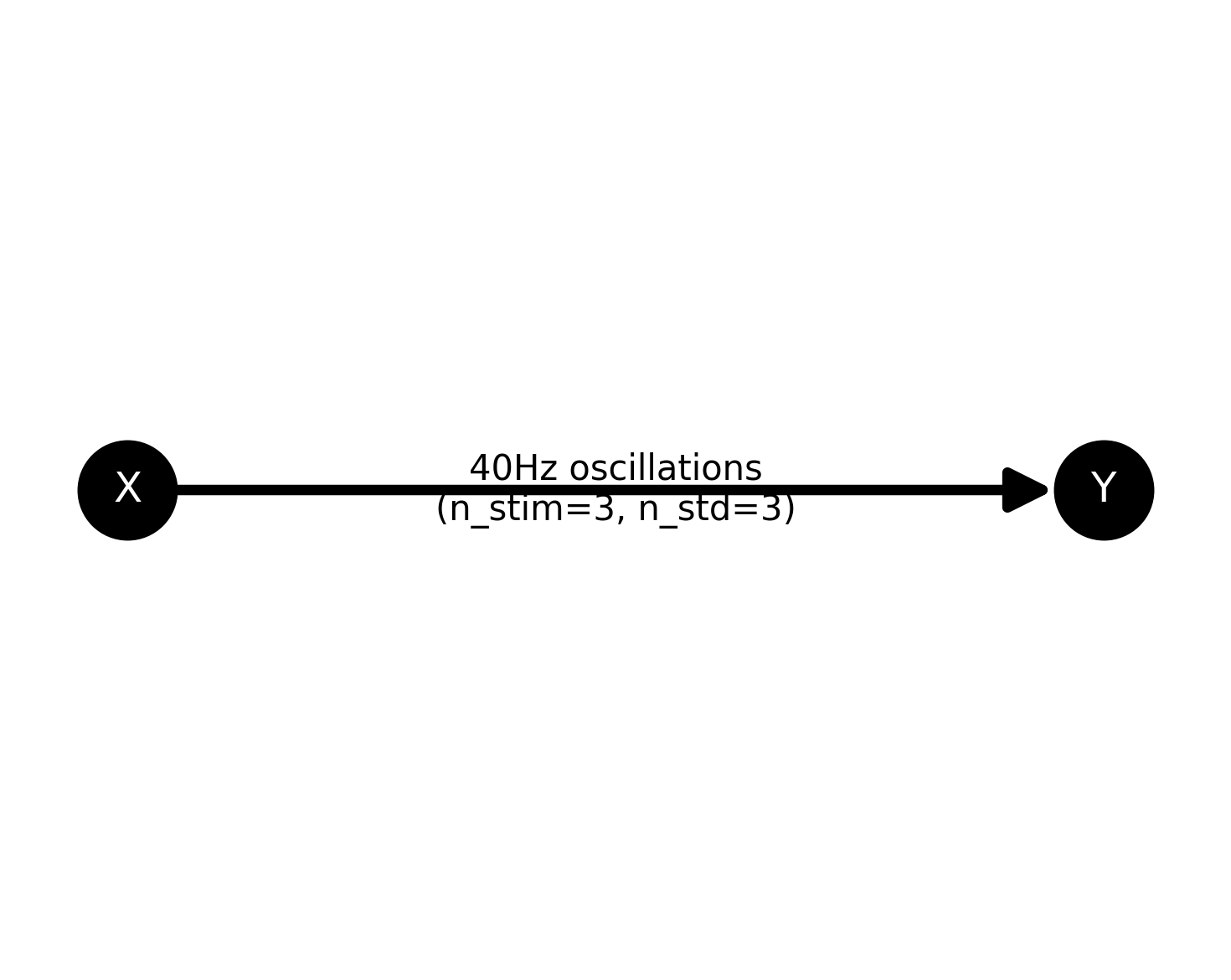
plot the network
plt.figure(figsize=(5, 4))
ss.plot_model()
plt.show()
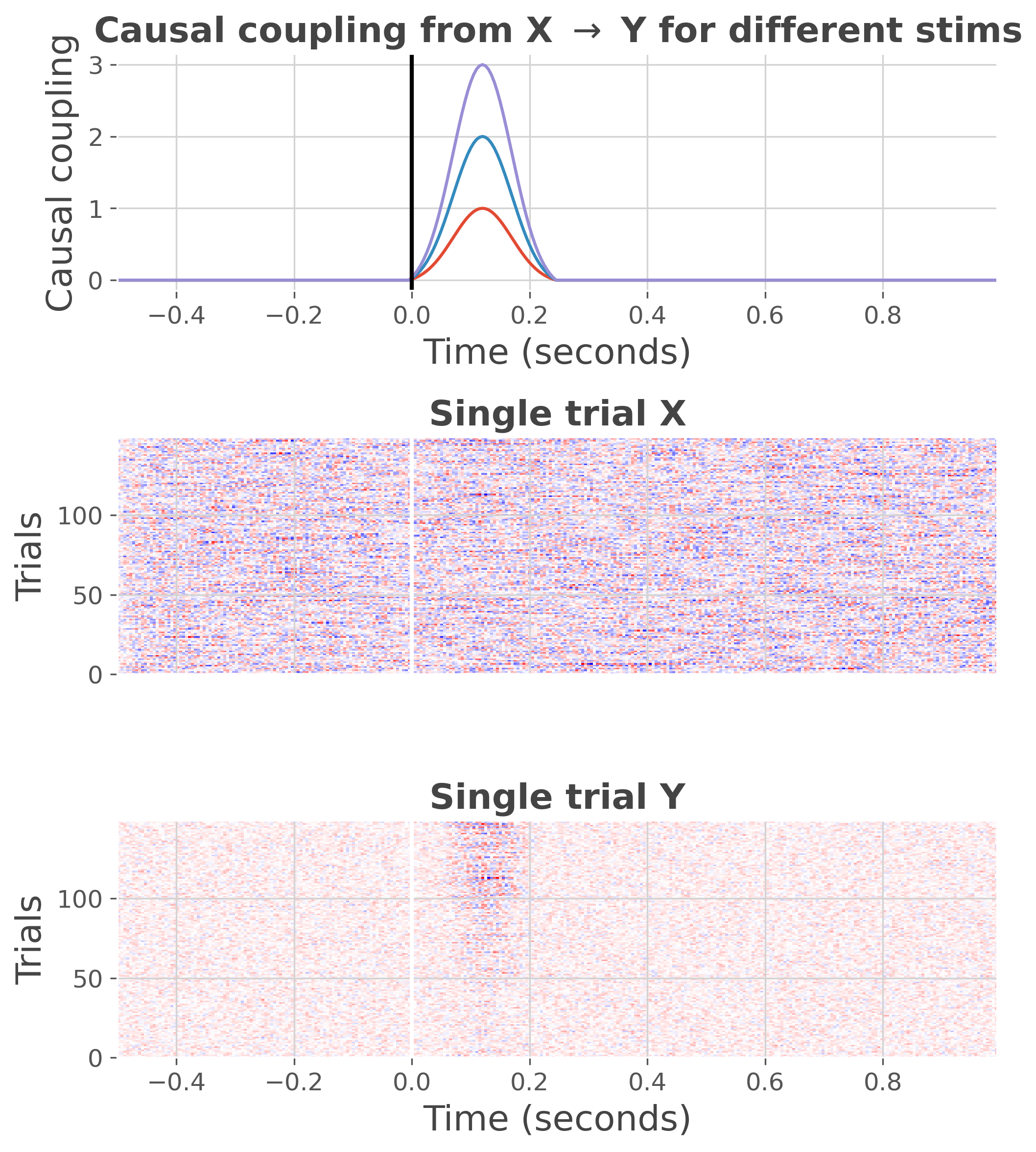
plot the data
plt.figure(figsize=(7, 8))
ss.plot(cmap='bwr')
plt.tight_layout()
plt.show()
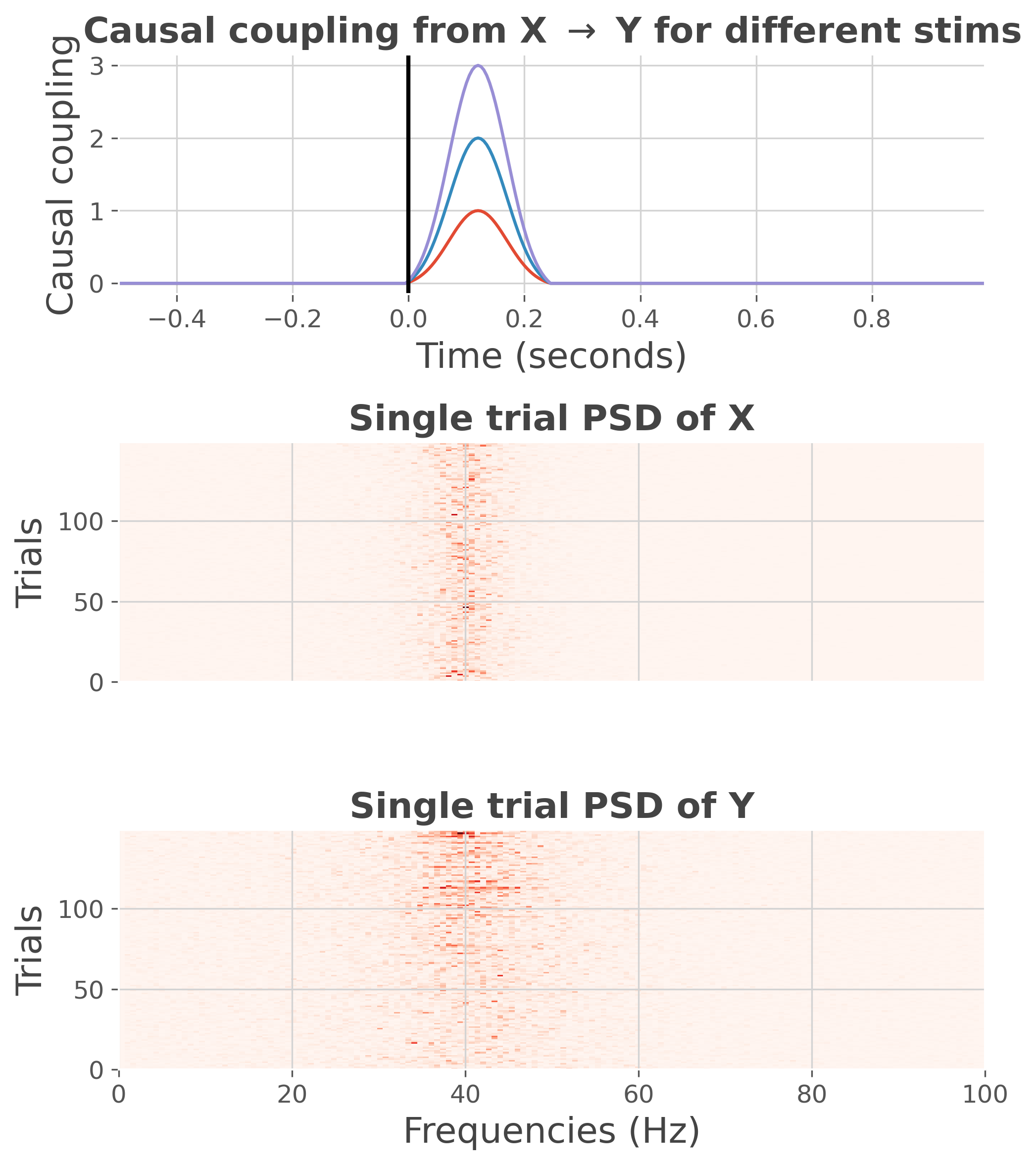
plot the power spectrum density (PSD)
plt.figure(figsize=(7, 8))
ss.plot(cmap='Reds', psd=True)
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 3.087 seconds)
Estimated memory usage: 449 MB