Note
Go to the end to download the full example code.
Compare within-subjects statistics when computing mutual information#
This example illustrates how to define and run a workflow for computing mutual information and evaluate stastitics. Inference are made within-subjects (fixed effect = ffx).
import numpy as np
from frites.simulations import sim_multi_suj_ephy, sim_mi_cc
from frites.dataset import DatasetEphy
from frites.workflow import WfMi
from frites import set_mpl_style
import matplotlib.pyplot as plt
set_mpl_style()
Simulate electrophysiological data#
Let’s start by simulating MEG / EEG electrophysiological data coming from
multiple subjects using the function
frites.simulations.sim_multi_suj_ephy(). As a result, the data output
is a list of length n_subjects of arrays, each one with a shape of
n_epochs, n_sites, n_times
modality = 'meeg'
n_subjects = 5
n_epochs = 400
n_times = 100
data, roi, time = sim_multi_suj_ephy(n_subjects=n_subjects, n_epochs=n_epochs,
n_times=n_times, modality=modality,
random_state=0)
Simulate mutual information#
Once the data have been created, we simulate an increase of mutual
information by creating a continuous variable y using the function
frites.simulations.sim_mi_cc(). This allows to simulate model-based
analysis by computing $I(data; y)$ where data and y are two continuous
variables
y, _ = sim_mi_cc(data, snr=.1)
Create an electrophysiological dataset#
Now, we use the frites.dataset.DatasetEphy in order to create a
compatible electrophysiological dataset
dt = DatasetEphy(data, y=y, roi=roi, times=time, verbose=False)
Define the workflow#
We now define the workflow for computing mi and evaluate statistics using the
class frites.workflow.WfMi. Here, the type of mutual
information to perform is ‘cc’ between it’s computed between two continuous
variables. And we also specify the inference type ‘ffx’ for fixed-effect
Compute the mutual information and statistics#
# list of corrections for multiple comparison
mcps = ['cluster', 'maxstat', 'fdr', 'bonferroni']
kw = dict(n_jobs=1, n_perm=200)
"""
The `cluster_th` input parameter specifies how the threshold is defined.
Use either :
* a float for a manual threshold
* None and it will be infered using the distribution of permutations
* 'tfce' for a TFCE threshold
"""
cluster_th = 'tfce' # {float, None, 'tfce'}
plt.figure(figsize=(10, 8))
for mcp in mcps:
print(f'-> FFX / MCP={mcp}')
mi, pvalues = wf.fit(dt, mcp=mcp, cluster_th=cluster_th, **kw)
# remove p-values that exceed 0.05
pvalues.data[pvalues >= .05] = 1.
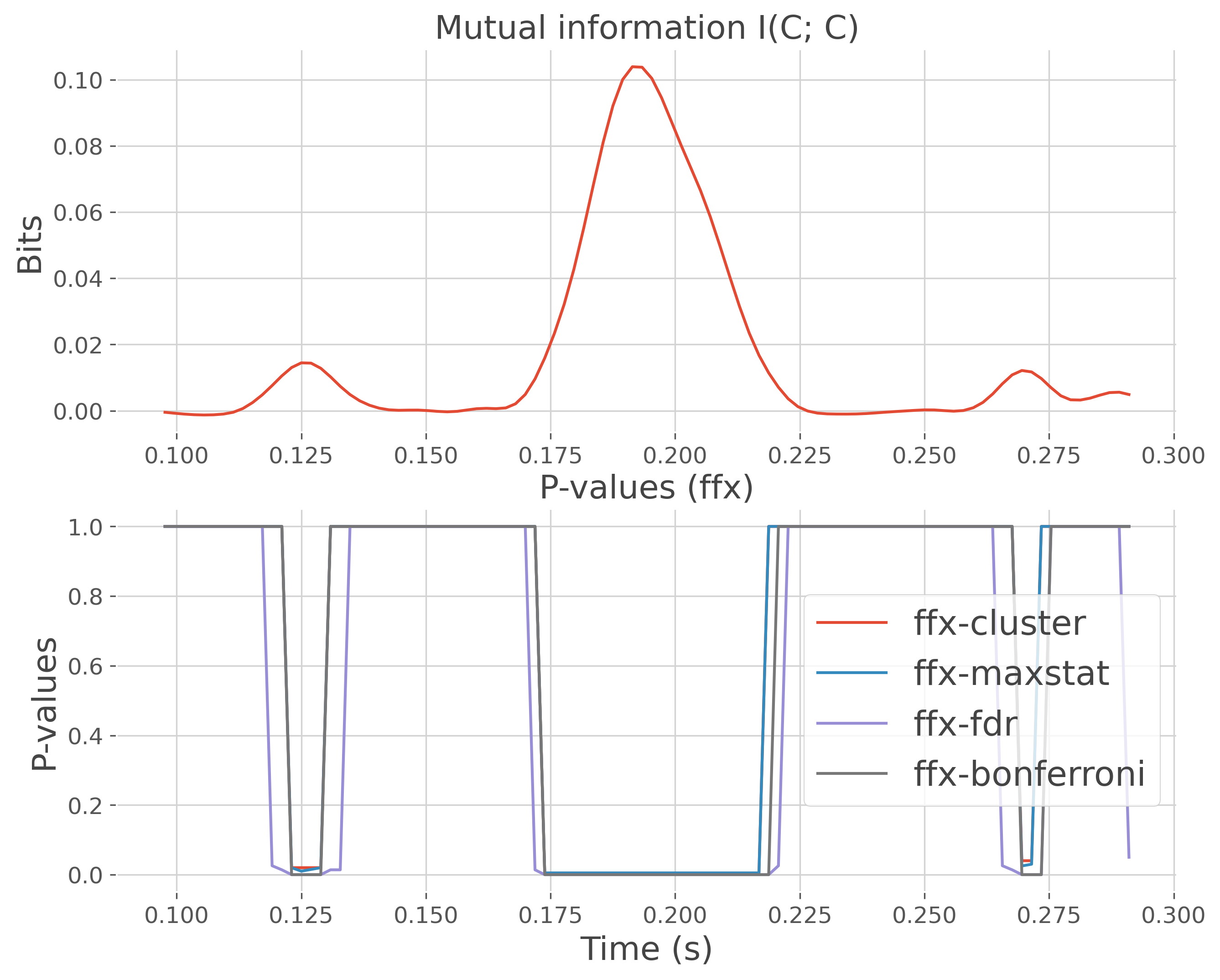
plt.subplot(212)
plt.plot(time, pvalues.squeeze(), label=f'ffx-{mcp}')
plt.xlabel('Time (s)'), plt.ylabel("P-values")
plt.title("P-values (ffx)")
plt.legend()
plt.subplot(211)
plt.plot(time, mi.squeeze())
plt.ylabel("Bits")
plt.title("Mutual information I(C; C)")
plt.show()
-> FFX / MCP=cluster
-> FFX / MCP=maxstat
-> FFX / MCP=fdr
-> FFX / MCP=bonferroni
Total running time of the script: (0 minutes 3.199 seconds)
Estimated memory usage: 395 MB